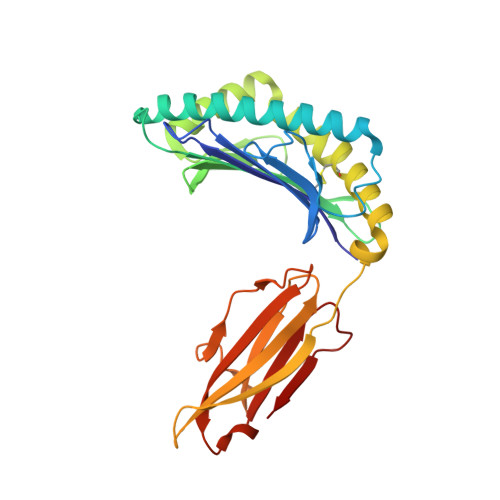
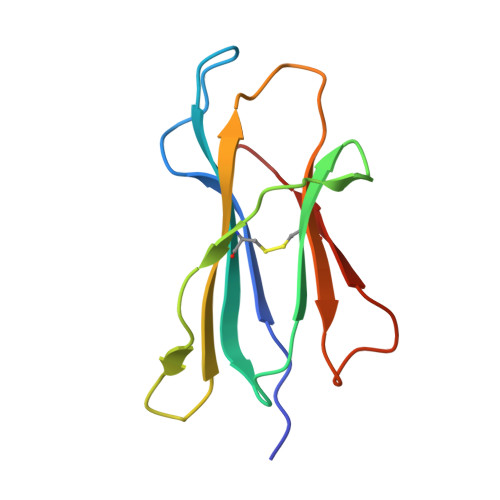
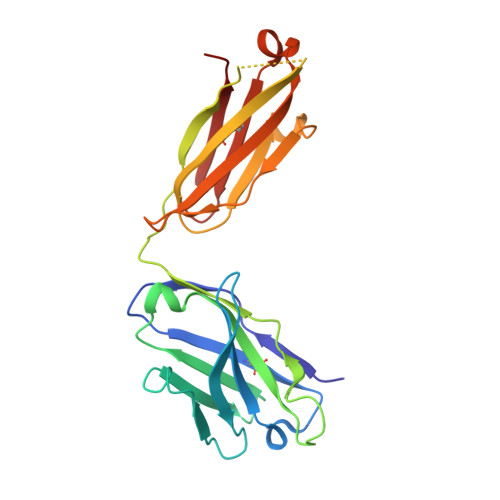
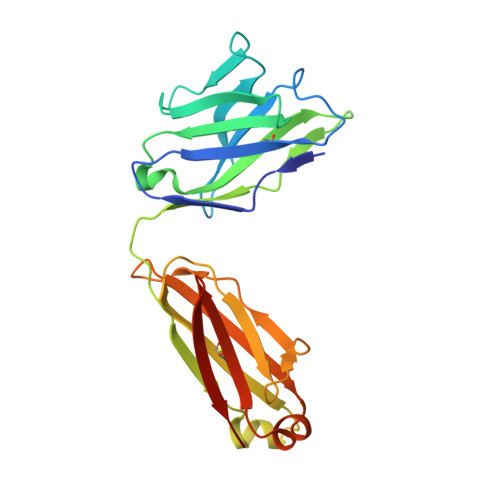
Defining the structural basis for human alloantibody binding to human leukocyte antigen allele HLA-A*11:01.
Gu, Y., Wong, Y.H., Liew, C.W., Chan, C.E.Z., Murali, T.M., Yap, J., Too, C.T., Purushotorman, K., Hamidinia, M., El Sahili, A., Goh, A.T.H., Teo, R.Z.C., Wood, K.J., Hanson, B.J., Gascoigne, N.R.J., Lescar, J., Vathsala, A., MacAry, P.A.(2019) Nat Commun 10: 893-893
- PubMed: 30792391
- DOI: https://doi.org/10.1038/s41467-019-08790-1
- Primary Citation of Related Structures:
6ID4 - PubMed Abstract:
Our understanding of the conformational and electrostatic determinants that underlie targeting of human leukocyte antigens (HLA) by anti-HLA alloantibodies is principally based upon in silico modelling. Here we provide a biochemical/biophysical and functional characterization of a human monoclonal alloantibody specific for a common HLA type, HLA-A*11:01. We present a 2.4 Å resolution map of the binding interface of this antibody on HLA-A*11:01 and compare the structural determinants with those utilized by T-cell receptor (TCR), killer-cell immunoglobulin-like receptor (KIR) and CD8 on the same molecule. These data provide a mechanistic insight into the paratope-epitope relationship between an alloantibody and its target HLA molecule in a biological context where other immune receptors are concomitantly engaged. This has important implications for our interpretation of serologic binding patterns of anti-HLA antibodies in sensitized individuals and thus, for the biology of human alloresponses.
- Department of Microbiology and Immunology, Yong Loo Lin School of Medicine, National University of Singapore, Singapore, Singapore.
Organizational Affiliation: