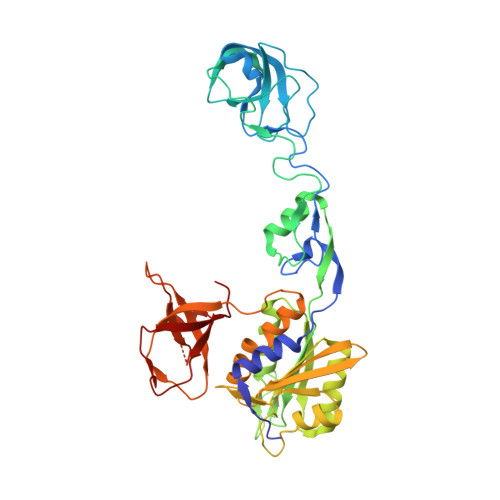
Elucidating the Molecular Basis for Inhibitory Neurotransmission Regulation by Artemisinins.
Kasaragod, V.B., Hausrat, T.J., Schaefer, N., Kuhn, M., Christensen, N.R., Tessmer, I., Maric, H.M., Madsen, K.L., Sotriffer, C., Villmann, C., Kneussel, M., Schindelin, H.(2019) Neuron 101: 673-689.e11
- PubMed: 30704910
- DOI: https://doi.org/10.1016/j.neuron.2019.01.001
- Primary Citation of Related Structures:
6FGC, 6FGD, 6HSN, 6HSO - PubMed Abstract:
The frontline anti-malarial drug artemisinin and its derivatives have also been implicated in modulating multiple mammalian cellular pathways, including the recent identification of targeting γ-aminobutyric acid type A receptor (GABA A R) signaling in the pancreas. Their molecular mechanism of action, however, remains elusive. Here, we present crystal structures of gephyrin, the central organizer at inhibitory postsynapses, in complex with artesunate and artemether at 1.5-Å resolution. These artemisinins target the universal inhibitory neurotransmitter receptor-binding epitope of gephyrin, thus inhibiting critical interactions between gephyrin and glycine receptors (GlyRs) as well as GABA A Rs. Electrophysiological recordings reveal a significant inhibition of gephyrin-mediated neurotransmission by artemisinins. Furthermore, clustering analyses in primary neurons demonstrate a rapid inhibition and a time-dependent regulation of gephyrin and GABA A R cluster parameters. Our data not only provide a comprehensive model for artemisinin-mediated modulation of inhibitory neurotransmission but also establish artemisinins as potential lead compounds to pharmacologically interfere with this process.
- Institute of Structural Biology, Rudolf Virchow Center for Experimental Biomedicine, University of Würzburg, Josef-Schneider-Str. 2, 97080 Würzburg, Germany.
Organizational Affiliation: