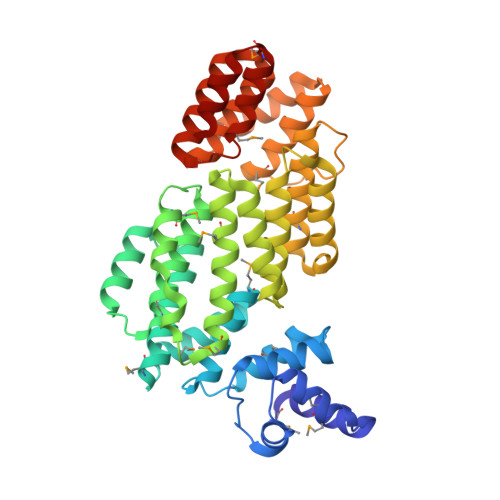
Deciphering the Molecular Mechanism Underpinning Phage Arbitrium Communication Systems.
Gallego Del Sol, F., Penades, J.R., Marina, A.(2019) Mol Cell 74: 59-72.e3
- PubMed: 30745087
- DOI: https://doi.org/10.1016/j.molcel.2019.01.025
- Primary Citation of Related Structures:
6HP3, 6HP5, 6HP7 - PubMed Abstract:
Bacillus phages use a communication system, termed "arbitrium," to coordinate lysis-lysogeny decisions. Arbitrium communication is mediated by the production and secretion of a hexapeptide (AimP) during lytic cycle. Once internalized, AimP reduces the expression of the negative regulator of lysogeny, AimX, by binding to the transcription factor, AimR, promoting lysogeny. We have elucidated the crystal structures of AimR from the Bacillus subtilis SPbeta phage in its apo form, bound to its DNA operator and in complex with AimP. AimR presents intrinsic plasticity, sharing structural features with the RRNPP quorum-sensing family. Remarkably, AimR binds to an unusual operator with a long spacer that interacts nonspecifically with the receptor TPR domain, while the HTH domain canonically recognizes two inverted repeats. AimP stabilizes a compact conformation of AimR that approximates the DNA-recognition helices, preventing AimR binding to the aimX promoter region. Our results establish the molecular basis of the arbitrium communication system.
- Instituto de Biomedicina de Valencia (IBV-CSIC) and CIBER de Enfermedades Raras (CIBERER), 46010 Valencia, Spain.
Organizational Affiliation: