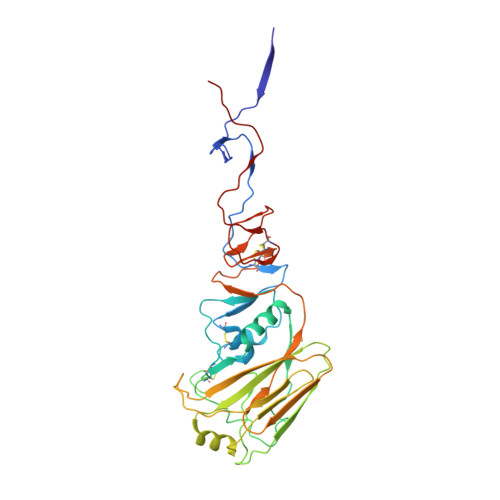
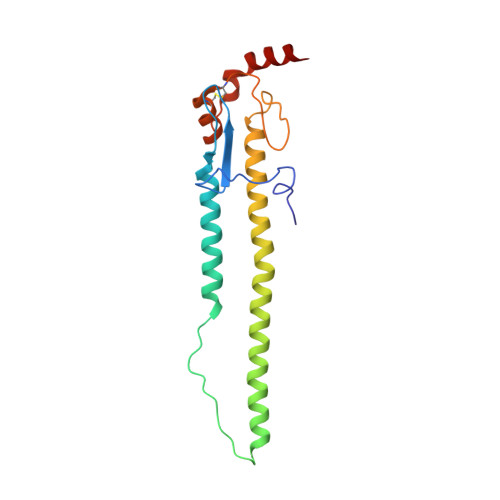
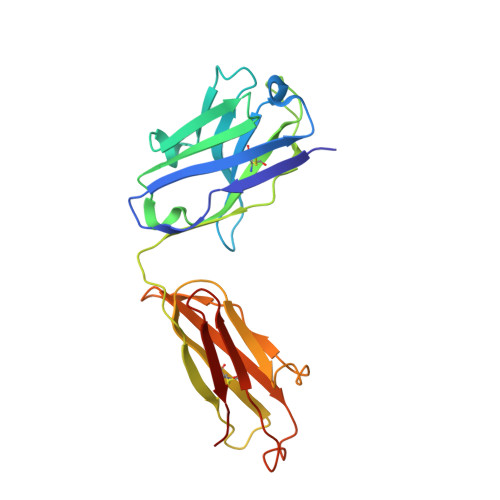
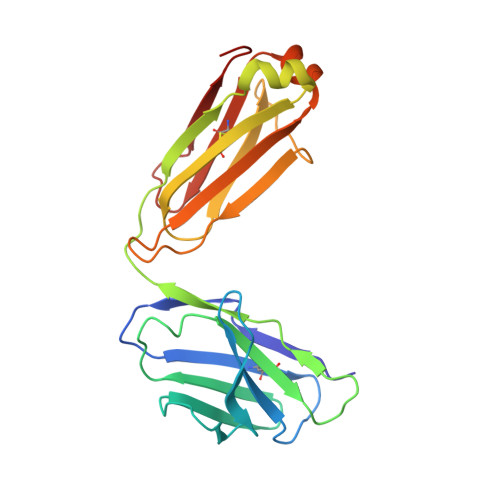
Influenza hemagglutinin membrane anchor.
Benton, D.J., Nans, A., Calder, L.J., Turner, J., Neu, U., Lin, Y.P., Ketelaars, E., Kallewaard, N.L., Corti, D., Lanzavecchia, A., Gamblin, S.J., Rosenthal, P.B., Skehel, J.J.(2018) Proc Natl Acad Sci U S A 115: 10112-10117
- PubMed: 30224494
- DOI: https://doi.org/10.1073/pnas.1810927115
- Primary Citation of Related Structures:
6HJN, 6HJP, 6HJQ, 6HJR, 6HKG - PubMed Abstract:
Viruses with membranes fuse them with cellular membranes, to transfer their genomes into cells at the beginning of infection. For Influenza virus, the membrane glycoprotein involved in fusion is the hemagglutinin (HA), the 3D structure of which is known from X-ray crystallographic studies. The soluble ectodomain fragments used in these studies lacked the "membrane anchor" portion of the molecule. Since this region has a role in membrane fusion, we have determined its structure by analyzing the intact, full-length molecule in a detergent micelle, using cryo-EM. We have also compared the structures of full-length HA-detergent micelles with full-length HA-Fab complex detergent micelles, to describe an infectivity-neutralizing monoclonal Fab that binds near the ectodomain membrane anchor junction. We determine a high-resolution HA structure which compares favorably in detail with the structure of the ectodomain seen by X-ray crystallography; we detect, clearly, all five carbohydrate side chains of HA; and we find that the ectodomain is joined to the membrane anchor by flexible, eight-residue-long, linkers. The linkers extend into the detergent micelle to join a central triple-helical structure that is a major component of the membrane anchor.
- Structural Biology of Disease Processes Laboratory, Francis Crick Institute, NW1 1AT London, United Kingdom; Donald.Benton@crick.ac.uk Peter.Rosenthal@crick.ac.uk John.Skehel@crick.ac.uk.
Organizational Affiliation: