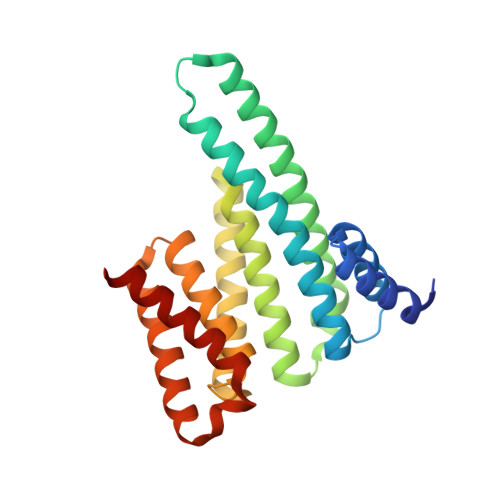
A Thermodynamic Model for Multivalency in 14-3-3 Protein-Protein Interactions.
Stevers, L.M., de Vink, P.J., Ottmann, C., Huskens, J., Brunsveld, L.(2018) J Am Chem Soc 140: 14498-14510
- PubMed: 30296824
- DOI: https://doi.org/10.1021/jacs.8b09618
- Primary Citation of Related Structures:
6HEP - PubMed Abstract:
Protein-protein interactions (PPIs) are at the core of molecular control over cellular function. Multivalency in PPI formation, such as via proteins with multiple binding sites and different valencies, requires fundamental understanding to address correlated challenges in pathologies and drug development. Thermodynamic binding models are needed to provide frameworks for describing multivalent PPIs. We established a model based on ditopic host-guest systems featuring the effective molarity, a hallmark property of multivalency, as a prime parameter governing the intramolecular binding in divalent interactions. By way of illustration, we study the interaction of the bivalent 14-3-3 protein scaffold with both the nonavalent CFTR and the hexavalent LRRK2 proteins, determining the underlying thermodynamics and providing insights into the role of individual sites in the context of the multivalent platform. Fitting of binding data reveals enthalpy-entropy correlation in both systems. Simulations of speciations for the entire phosphorylated protein domains reveal that the CFTR protein preferably binds to 14-3-3 by combinations including the strongest binding site pS768, but that other binding sites take over when this site is eliminated, leading to only a minor decrease in total affinity for 14-3-3. For LRRK2, two binding sites dominate the complex formation with 14-3-3, but the distantly located pS1444 site also plays a role in complex formation. Thermodynamic modeling of these multivalent PPIs allowed analyzing and predicting the effects of individual sites regarding their modulation via, for example, (de)phosphorylation or small-molecule targeting. The results specifically bring forward the potential of PPI stabilization, as an entry for drug discovery for multivalent PPIs.
- Laboratory of Chemical Biology, Department of Biomedical Engineering and Institute for Complex Molecular Systems , Technische Universiteit Eindhoven , P.O. Box 513, Eindhoven 5600 MB , The Netherlands.
Organizational Affiliation: