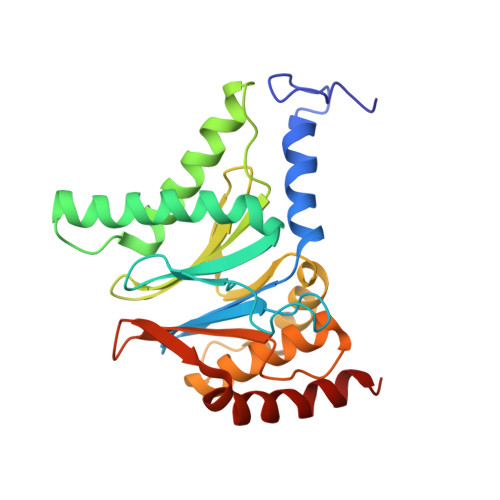
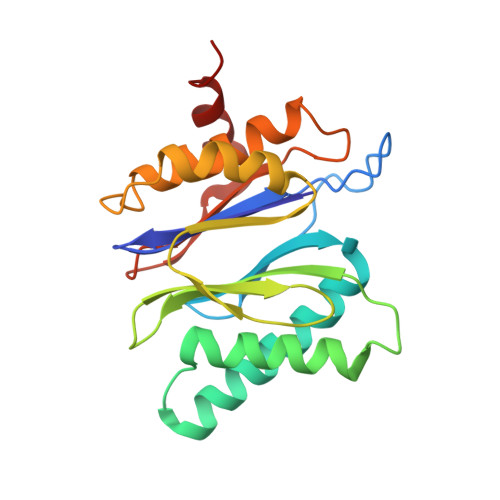
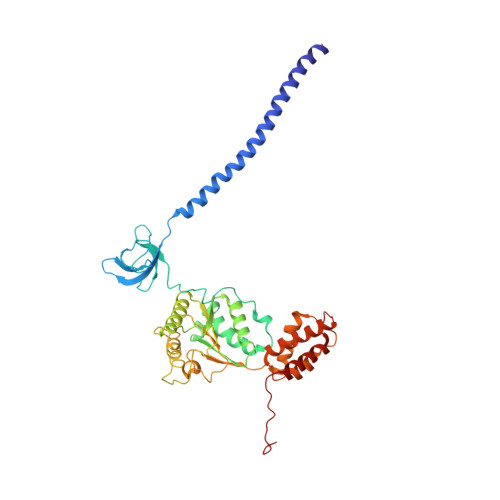
Cryo-EM structures of the archaeal PAN-proteasome reveal an around-the-ring ATPase cycle.
Majumder, P., Rudack, T., Beck, F., Danev, R., Pfeifer, G., Nagy, I., Baumeister, W.(2019) Proc Natl Acad Sci U S A 116: 534-539
- PubMed: 30559193
- DOI: https://doi.org/10.1073/pnas.1817752116
- Primary Citation of Related Structures:
6HE4, 6HE5, 6HE7, 6HE8, 6HE9, 6HEA, 6HEC, 6HED - PubMed Abstract:
Proteasomes occur in all three domains of life, and are the principal molecular machines for the regulated degradation of intracellular proteins. They play key roles in the maintenance of protein homeostasis, and control vital cellular processes. While the eukaryotic 26S proteasome is extensively characterized, its putative evolutionary precursor, the archaeal proteasome, remains poorly understood. The primordial archaeal proteasome consists of a 20S proteolytic core particle (CP), and an AAA-ATPase module. This minimal complex degrades protein unassisted by non-ATPase subunits that are present in a 26S proteasome regulatory particle (RP). Using cryo-EM single-particle analysis, we determined structures of the archaeal CP in complex with the AAA-ATPase PAN (proteasome-activating nucleotidase). Five conformational states were identified, elucidating the functional cycle of PAN, and its interaction with the CP. Coexisting nucleotide states, and correlated intersubunit signaling features, coordinate rotation of the PAN-ATPase staircase, and allosterically regulate N-domain motions and CP gate opening. These findings reveal the structural basis for a sequential around-the-ring ATPase cycle, which is likely conserved in AAA-ATPases.
- Department of Molecular Structural Biology, Max Planck Institute of Biochemistry, 82152 Martinsried, Germany.
Organizational Affiliation: