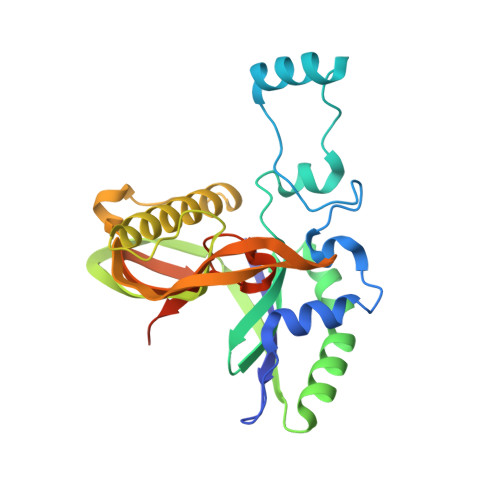
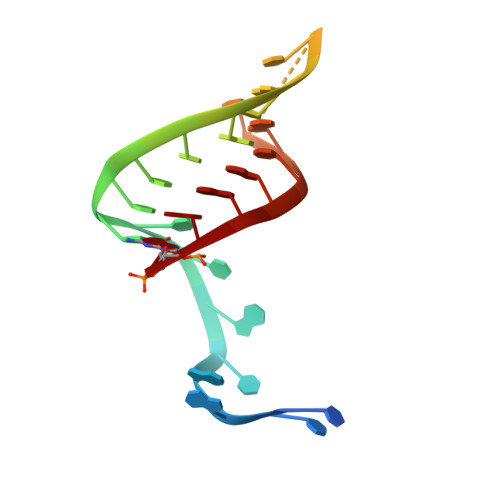
Type IV CRISPR RNA processing and effector complex formation in Aromatoleum aromaticum.
Ozcan, A., Pausch, P., Linden, A., Wulf, A., Schuhle, K., Heider, J., Urlaub, H., Heimerl, T., Bange, G., Randau, L.(2019) Nat Microbiol 4: 89-96
- PubMed: 30397343
- DOI: https://doi.org/10.1038/s41564-018-0274-8
- Primary Citation of Related Structures:
6H9H, 6H9I - PubMed Abstract:
Type IV CRISPR-Cas modules belong to class 1 prokaryotic adaptive immune systems, which are defined by the presence of multisubunit effector complexes. They usually lack the known Cas proteins involved in adaptation and target cleavage, and their function has not been experimentally addressed. To investigate RNA and protein components of this CRISPR-Cas type, we located a complete type IV cas gene locus and an adjacent CRISPR array on a megaplasmid of Aromatoleum aromaticum EbN1, which contains an additional type I-C system on its chromosome. RNA sequencing analyses verified CRISPR RNA (crRNA) production and maturation for both systems. Type IV crRNAs were shown to harbour unusually short 7 nucleotide 5'-repeat tags and stable 3' hairpin structures. A unique Cas6 variant (Csf5) was identified that generates crRNAs that are specifically incorporated into type IV CRISPR-ribonucleoprotein (crRNP) complexes. Structures of RNA-bound Csf5 were obtained. Recombinant production and purification of the type IV Cas proteins, together with electron microscopy, revealed that Csf2 acts as a helical backbone for type IV crRNPs that include Csf5, Csf3 and a large subunit (Csf1). Mass spectrometry analyses identified protein-protein and protein-RNA contact sites. These results highlight evolutionary connections between type IV and type I CRISPR-Cas systems and demonstrate that type IV CRISPR-Cas systems employ crRNA-guided effector complexes.
- Max Planck Institute for Terrestrial Microbiology, Marburg, Germany.
Organizational Affiliation: