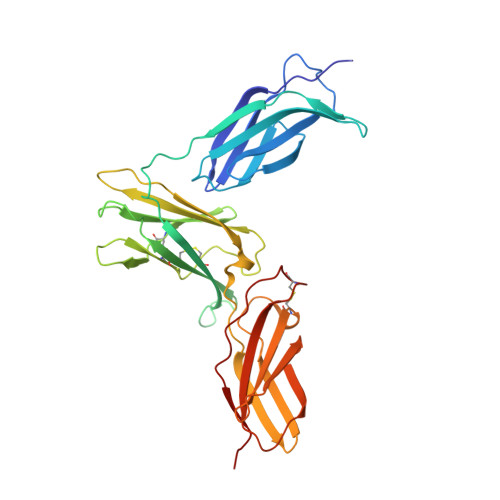
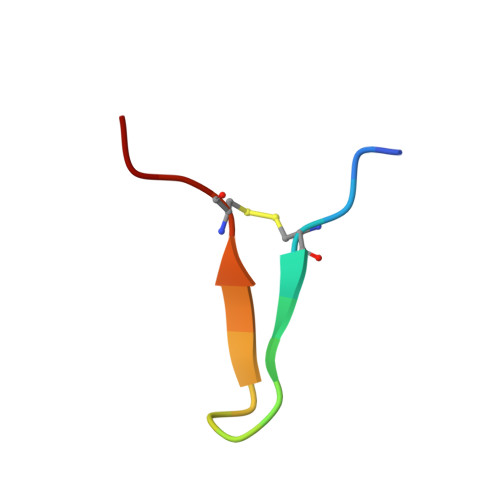
Structural Basis of Interleukin-5 Inhibition by the Small Cyclic Peptide AF17121.
Scheide-Noeth, J.P., Rosen, M., Baumstark, D., Dietz, H., Mueller, T.D.(2019) J Mol Biology 431: 714-731
- PubMed: 30529748
- DOI: https://doi.org/10.1016/j.jmb.2018.11.029
- Primary Citation of Related Structures:
6H41 - PubMed Abstract:
Interleukin-5 (IL-5) is a T-helper cell of subtype 2 cytokine involved in many aspects of eosinophil life. Eosinophilic granulocytes play a pathogenic role in the progression of atopic diseases, such as allergy, asthma and atopic dermatitis and hypereosinophilic syndromes. Here, eosinophils upon activation degranulate leading to the release of proinflammatory proteins and mediators stored in intracellular vesicles termed granula thereby causing local inflammation, which when persisting leads to tissue damage and organ failure. As a key regulator of eosinophil function, IL-5 therefore presents a major pharmaceutical target and approaches to interfere with IL-5 receptor activation are of great interest. Here we present the structure of the IL-5 inhibiting peptide AF17121 bound to the extracellular domain of the IL-5 receptor IL-5Rα. The small 18mer cyclic peptide snugly fits into the wrench-like cleft of the IL-5 receptor, thereby blocking access of key residues for IL-5 binding. While AF17121 and IL-5 seemingly bind to a similar epitope at IL-5Rα, functional studies show that recognition and binding of both ligands differ. Using the structure data, peptide variants with improved IL-5 inhibition have been generated, which might present valuable starting points for superior peptide-based IL-5 antagonists.
- Department of Molecular Plant Physiology and Biophysics, Julius-von-Sachs Institute of the University Wuerzburg, Julius-von-Sachs-Platz 2, D-97082, Wuerzburg, Germany.
Organizational Affiliation: