A Giant Extracellular Matrix Binding Protein of Staphylococcus epidermidis Binds Surface-Immobilized Fibronectin via a Novel Mechanism.
Buttner, H., Perbandt, M., Kohler, T., Kikhney, A., Wolters, M., Christner, M., Heise, M., Wilde, J., Weisselberg, S., Both, A., Betzel, C., Hammerschmidt, S., Svergun, D., Aepfelbacher, M., Rohde, H.(2020) mBio 11
- PubMed: 33082256
- DOI: https://doi.org/10.1128/mBio.01612-20
- Primary Citation of Related Structures:
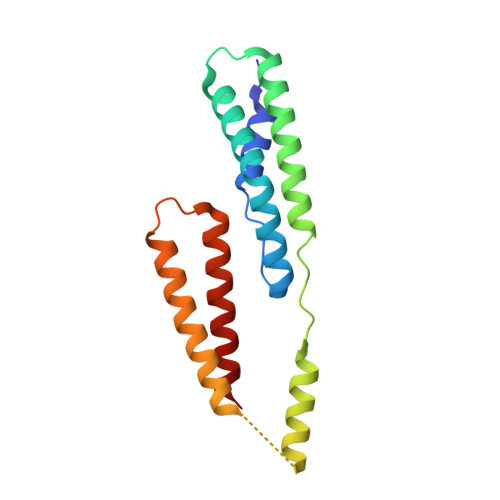
6GV5, 6GV8 - PubMed Abstract:
Although it is normally an innocuous part of the human skin microbiota, Staphylococcus epidermidis has emerged as a major nosocomial pathogen, and implanted foreign materials are an essential risk factor for the development of an infection. The extraordinary efficiency of S. epidermidis to colonize artificial surfaces is particularly related to the ability to form biofilms. Biofilm formation itself critically depends on stable pathogen binding to extracellular host matrix components, e.g. fibronectin (Fn), covering inserted devices in vast amounts. Extracellular matrix binding protein (Embp) and its subdomains referred to as the F-repeat and the FG-repeat are critical for adherence of S. epidermidis to surface-immobilized Fn. Embp-Fn interactions preferentially occur with surface-bound, but not folded, globular Fn via binding to the F3 domain. High-resolution structure analysis of F- and FG-repeats revealed that both repeats are composed of two tightly connected triple α-helix bundles, exhibiting an elongated but rather rigid structural organization in solution. Both F- and FG-repeat possess Fn-binding capacity via interactions with type III subdomain FN12, involving residues within the C and F β-sheet. FN12 essentially supports stability of the globular Fn state, and thus these findings reasonably explain why Embp-mediated interaction of S. epidermidis necessitates Fn surface immobilization. Thus, Embp employs an uncharacterized bacterial Fn-binding mechanism to promote staphylococcal adherence. IMPORTANCE Staphylococcus epidermidis is a leading pathogen in implant-associated hospital infections. The pathogenesis critically depends on bacterial binding to ECM components, specifically fibronectin (Fn). The cell surface-localized, 1-MDa extracellular matrix binding protein (Embp) is essentially characterized by 10 F- and 40 FG-repeats. These repetitive units, each characterized by two α-helical bundles, organize themselves in a rigid, elongated form. Embp binds preferentially to surface-localized but not soluble Fn, with both F- and FG-repeats being sufficient for Fn binding and resulting bacterial adherence. Binding preferentially involves Fn type III domain, specifically residues of FN12 β-sheets C and F. Both play key role in stabilizing the globular Fn conformation, explaining the necessity of Fn surface immobilization for a subsequent interaction with Embp. In comparison to many other bacterial Fn-binding proteins using the Fn N terminus, Embp employs a previously undescribed mechanism supporting the adhesion of S. epidermidis to surface-immobilized Fn.
- Institut for Medical Microbiology, Virology, and Hygiene, University Medical Center Hamburg-Eppendorf, Hamburg-Eppendorf, Germany.
Organizational Affiliation: