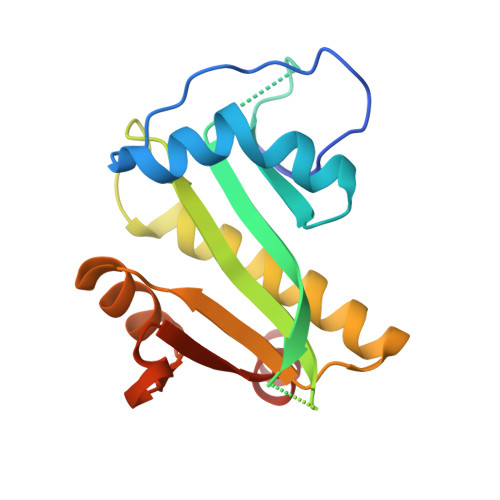
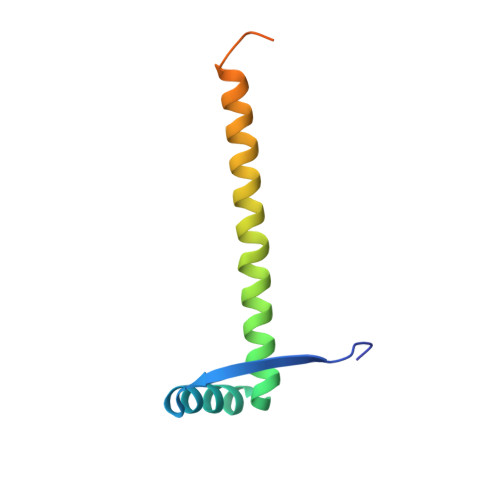
Mechanism of regulation and neutralization of the AtaR-AtaT toxin-antitoxin system.
Jurenas, D., Van Melderen, L., Garcia-Pino, A.(2019) Nat Chem Biol 15: 285-294
- PubMed: 30718814
- DOI: https://doi.org/10.1038/s41589-018-0216-z
- Primary Citation of Related Structures:
6GTO, 6GTP, 6GTQ, 6GTR, 6GTS - PubMed Abstract:
GCN5-related N-acetyl-transferase (GNAT)-like enzymes from toxin-antitoxin modules are strong inhibitors of protein synthesis. Here, we present the bases of the regulatory mechanisms of ataRT, a model GNAT-toxin-antitoxin module, from toxin synthesis to its action as a transcriptional de-repressor. We show the antitoxin (AtaR) traps the toxin (AtaT) in a pre-catalytic monomeric state and precludes the effective binding of ac-CoA and its target Met-transfer RNA fMet . In the repressor complex, AtaR intrinsically disordered region interacts with AtaT at two different sites, folding into different structures, that are involved in two separate functional roles, toxin neutralization and placing the DNA-binding domains of AtaR in a binding-compatible orientation. Our data suggests AtaR neutralizes AtaT as a monomer, right after its synthesis and only the toxin-antitoxin complex formed in this way is an active repressor. Once activated by dimerization, later neutralization of the toxin results in a toxin-antitoxin complex that is not able to repress transcription.
- Cellular and Molecular Microbiology, Department of Molecular Biology, Université Libre de Bruxelles, Gosselies, Belgium.
Organizational Affiliation: