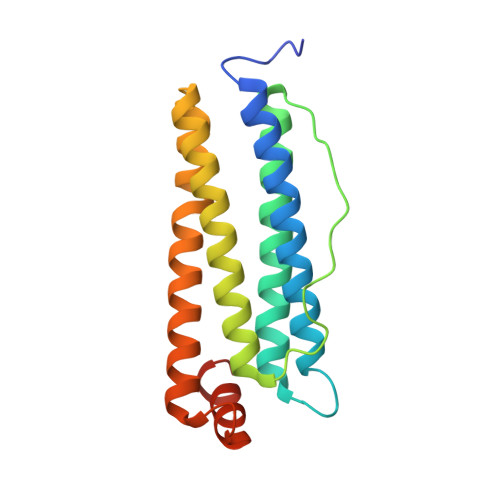
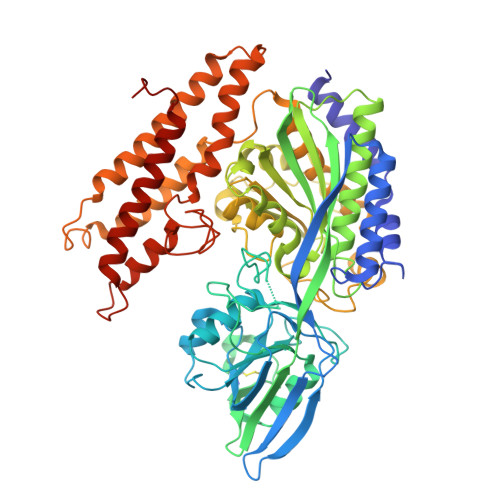
Cryo-EM structure of the human ferritin-transferrin receptor 1 complex.
Montemiglio, L.C., Testi, C., Ceci, P., Falvo, E., Pitea, M., Savino, C., Arcovito, A., Peruzzi, G., Baiocco, P., Mancia, F., Boffi, A., des Georges, A., Vallone, B.(2019) Nat Commun 10: 1121-1121
- PubMed: 30850661
- DOI: https://doi.org/10.1038/s41467-019-09098-w
- Primary Citation of Related Structures:
6GSR, 6H5I - PubMed Abstract:
Human transferrin receptor 1 (CD71) guarantees iron supply by endocytosis upon binding of iron-loaded transferrin and ferritin. Arenaviruses and the malaria parasite exploit CD71 for cell invasion and epitopes on CD71 for interaction with transferrin and pathogenic hosts were identified. Here, we provide the molecular basis of the CD71 ectodomain-human ferritin interaction by determining the 3.9 Å resolution single-particle cryo-electron microscopy structure of their complex and by validating our structural findings in a cellular context. The contact surfaces between the heavy-chain ferritin and CD71 largely overlap with arenaviruses and Plasmodium vivax binding regions in the apical part of the receptor ectodomain. Our data account for transferrin-independent binding of ferritin to CD71 and suggest that select pathogens may have adapted to enter cells by mimicking the ferritin access gate.
- Department of Biochemical Sciences "Alessandro Rossi Fanelli", Sapienza University of Rome, P.le A. Moro 5, 00185, Rome, Italy.
Organizational Affiliation: