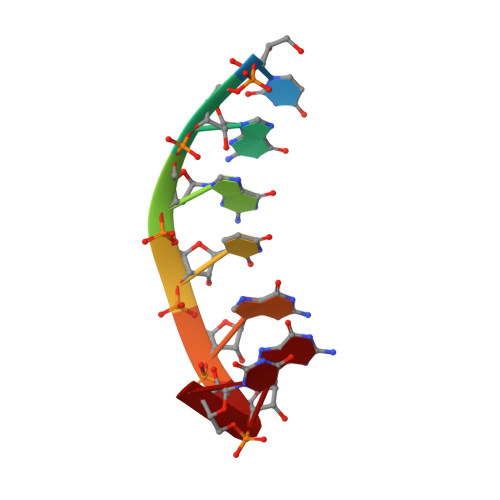
Unraveling the structural basis for the exceptional stability of RNA G-quadruplexes capped by a uridine tetrad at the 3' terminus.
Andralojc, W., Malgowska, M., Sarzynska, J., Pasternak, K., Szpotkowski, K., Kierzek, R., Gdaniec, Z.(2019) RNA 25: 121-134
- PubMed: 30341177
- DOI: https://doi.org/10.1261/rna.068163.118
- Primary Citation of Related Structures:
6GE1 - PubMed Abstract:
Uridine tetrads (U-tetrads) are a structural element encountered in RNA G-quadruplexes, for example, in the structures formed by the biologically relevant human telomeric repeat RNA. For these molecules, an unexpectedly strong stabilizing influence of a U-tetrad forming at the 3' terminus of a quadruplex was reported. Here we present the high-resolution solution NMR structure of the r(UGGUGGU) 4 quadruplex which, in our opinion, provides an explanation for this stabilization. Our structure features a distinctive, abrupt chain reversal just prior to the 3' uridine tetrad. Similar "reversed U-tetrads" were already observed in the crystalline phase. However, our NMR structure coupled with extensive explicit solvent molecular dynamics (MD) simulations identifies some key features of this motif that up to now remained overlooked. These include the presence of an exceptionally stable 2'OH to phosphate hydrogen bond, as well as the formation of an additional K + binding pocket in the quadruplex groove.
- Institute of Bioorganic Chemistry, Polish Academy of Sciences, 61-704 Poznan, Poland.
Organizational Affiliation: