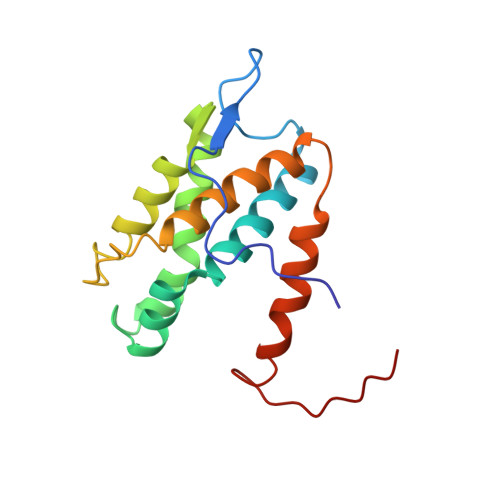
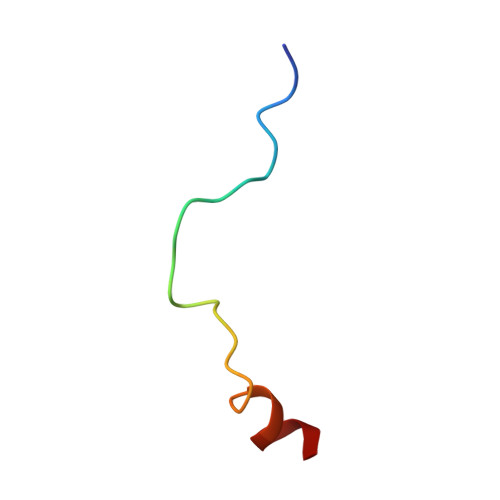
Molecular basis for disassembly of an importin:ribosomal protein complex by the escortin Tsr2.
Schutz, S., Michel, E., Damberger, F.F., Oplova, M., Pena, C., Leitner, A., Aebersold, R., Allain, F.H., Panse, V.G.(2018) Nat Commun 9: 3669-3669
- PubMed: 30201955
- DOI: https://doi.org/10.1038/s41467-018-06160-x
- Primary Citation of Related Structures:
6G03, 6G04 - PubMed Abstract:
Disordered extensions at the termini and short internal insertions distinguish eukaryotic ribosomal proteins (r-proteins) from their anucleated archaeal counterparts. Here, we report an NMR structure of such a eukaryotic-specific segment (ESS) in the r-protein eS26 in complex with the escortin Tsr2. The structure reveals how ESS attracts Tsr2 specifically to importin:eS26 complexes entering the nucleus in order to trigger non-canonical RanGTP-independent disassembly. Tsr2 then sequesters the released eS26 and prevents rebinding to the importin, providing an alternative allosteric mechanism to terminate the process of nuclear import. Notably, a Diamond-Blackfan anemia-associated Tsr2 mutant protein is impaired in binding to ESS, unveiling a critical role for this interaction in human hematopoiesis. We propose that eS26-ESS and Tsr2 are components of a nuclear sorting system that co-evolved with the emergence of the nucleocytoplasmic barrier and transport carriers.
- Institute of Medical Microbiology, University of Zurich, 8006, Zurich, Switzerland.
Organizational Affiliation: