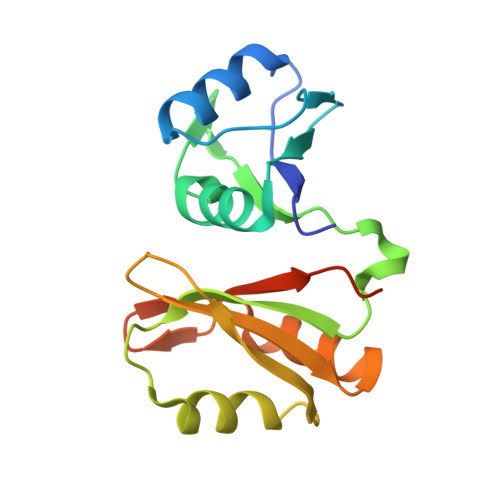
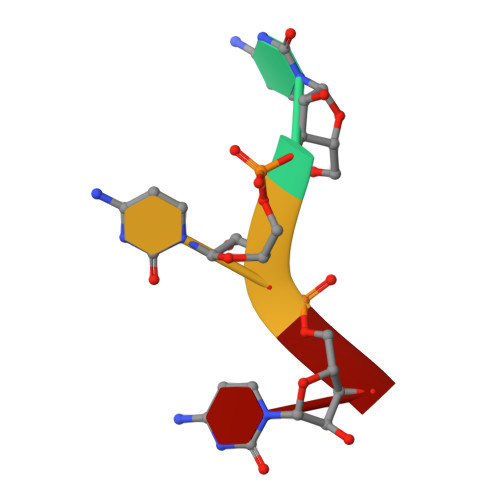
Structural basis of IMP3 RRM12 recognition of RNA.
Jia, M., Gut, H., Chao, J.A.(2018) RNA 24: 1659-1666
- PubMed: 30135093
- DOI: https://doi.org/10.1261/rna.065649.118
- Primary Citation of Related Structures:
6FQ1, 6FQR, 6GX6 - PubMed Abstract:
The IMP family of RNA binding proteins, also named as insulin-like growth factor 2 (IGF2) mRNA-binding proteins (IGF2BPs), are highly conserved RNA regulators that are involved in many RNA processing stages, including mRNA stability, localization, and translation. There are three paralogs in the IMP family, IMP1-3, in mammals that all adopt the same domain arrangement with two RNA recognition motifs (RRM) in the N terminus and four KH domains in the C terminus. Here, we report the structure and biochemical characterization of IMP3 RRM12 and its complex with two short RNAs. These structures show that both RRM domains of IMP3 adopt the canonical RRM topology with two α-helices packed on an anti-parallel four stranded β-sheet. The spatial orientation of RRM1 to RRM2 is unique compared with other known tandem RRM structures. In the IMP3 RRM12 complex with RNA, only RRM1 is involved in RNA binding and recognizes a dinucleotide sequence.
- Friedrich Miescher Institute for Biomedical Research, CH-4058 Basel, Switzerland.
Organizational Affiliation: