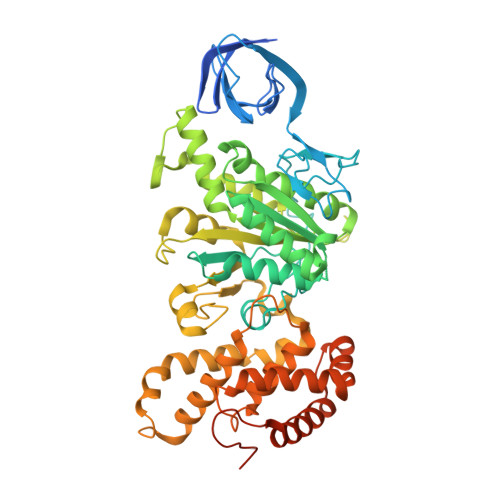
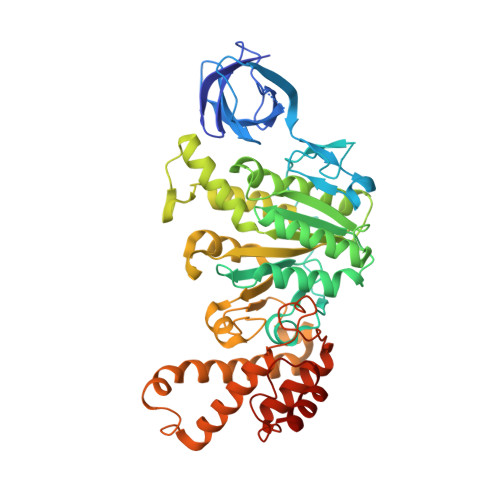
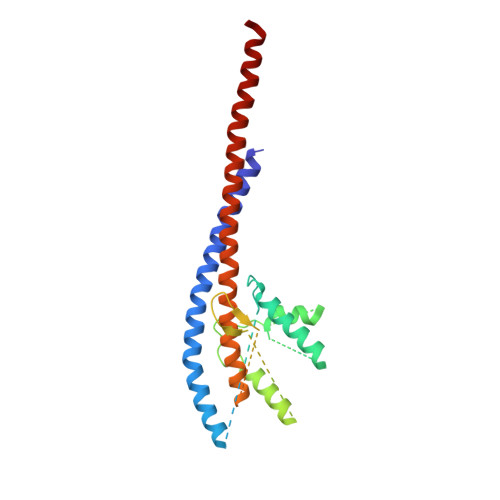
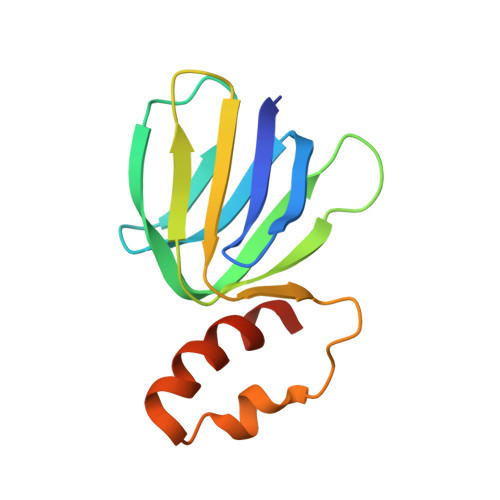
The structure of the catalytic domain of the ATP synthase fromMycobacterium smegmatisis a target for developing antitubercular drugs.
Zhang, A.T., Montgomery, M.G., Leslie, A.G.W., Cook, G.M., Walker, J.E.(2019) Proc Natl Acad Sci U S A 116: 4206-4211
- PubMed: 30683723
- DOI: https://doi.org/10.1073/pnas.1817615116
- Primary Citation of Related Structures:
6FOC - PubMed Abstract:
The crystal structure of the F 1 -catalytic domain of the adenosine triphosphate (ATP) synthase has been determined from Mycobacterium smegmatis which hydrolyzes ATP very poorly. The structure of the α 3 β 3 -component of the catalytic domain is similar to those in active F 1 -ATPases in Escherichia coli and Geobacillus stearothermophilus However, its ε-subunit differs from those in these two active bacterial F 1 -ATPases as an ATP molecule is not bound to the two α-helices forming its C-terminal domain, probably because they are shorter than those in active enzymes and they lack an amino acid that contributes to the ATP binding site in active enzymes. In E. coli and G. stearothermophilus , the α-helices adopt an "up" state where the α-helices enter the α 3 β 3 -domain and prevent the rotor from turning. The mycobacterial F 1 -ATPase is most similar to the F 1 -ATPase from Caldalkalibacillus thermarum , which also hydrolyzes ATP poorly. The β E -subunits in both enzymes are in the usual "open" conformation but appear to be occupied uniquely by the combination of an adenosine 5'-diphosphate molecule with no magnesium ion plus phosphate. This occupation is consistent with the finding that their rotors have been arrested at the same point in their rotary catalytic cycles. These bound hydrolytic products are probably the basis of the inhibition of ATP hydrolysis. It can be envisaged that specific as yet unidentified small molecules might bind to the F 1 domain in Mycobacterium tuberculosis , prevent ATP synthesis, and inhibit the growth of the pathogen.
- The Medical Research Council Mitochondrial Biology Unit, University of Cambridge, Cambridge Biomedical Campus, CB2 0XY Cambridge, United Kingdom.
Organizational Affiliation: