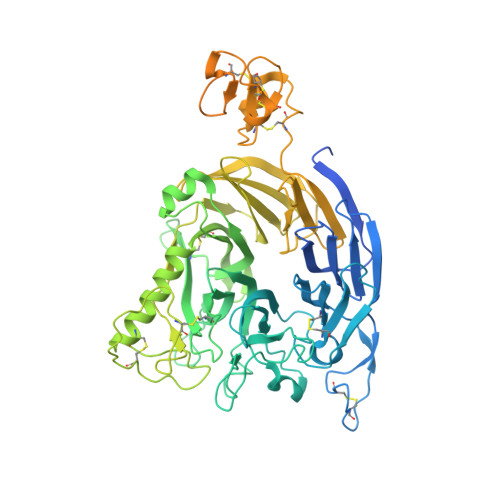
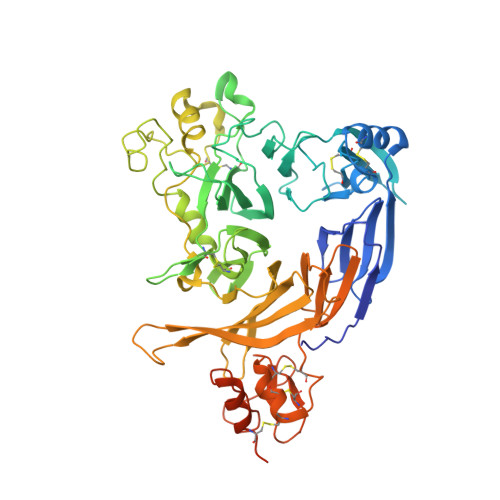
Structural basis of semaphorin-plexin cis interaction.
Rozbesky, D., Verhagen, M.G., Karia, D., Nagy, G.N., Alvarez, L., Robinson, R.A., Harlos, K., Padilla-Parra, S., Pasterkamp, R.J., Jones, E.Y.(2020) EMBO J : e102926-e102926
- PubMed: 32500924
- DOI: https://doi.org/10.15252/embj.2019102926
- Primary Citation of Related Structures:
6FKM, 6FKN - PubMed Abstract:
Semaphorin ligands interact with plexin receptors to contribute to functions in the development of myriad tissues including neurite guidance and synaptic organisation within the nervous system. Cell-attached semaphorins interact in trans with plexins on opposing cells, but also in cis on the same cell. The interplay between trans and cis interactions is crucial for the regulated development of complex neural circuitry, but the underlying molecular mechanisms are uncharacterised. We have discovered a distinct mode of interaction through which the Drosophila semaphorin Sema1b and mouse Sema6A mediate binding in cis to their cognate plexin receptors. Our high-resolution structural, biophysical and in vitro analyses demonstrate that monomeric semaphorins can mediate a distinctive plexin binding mode. These findings suggest the interplay between monomeric vs dimeric states has a hereto unappreciated role in semaphorin biology, providing a mechanism by which Sema6s may balance cis and trans functionalities.
- Division of Structural Biology, Wellcome Centre for Human Genetics, University of Oxford, Oxford, UK.
Organizational Affiliation: