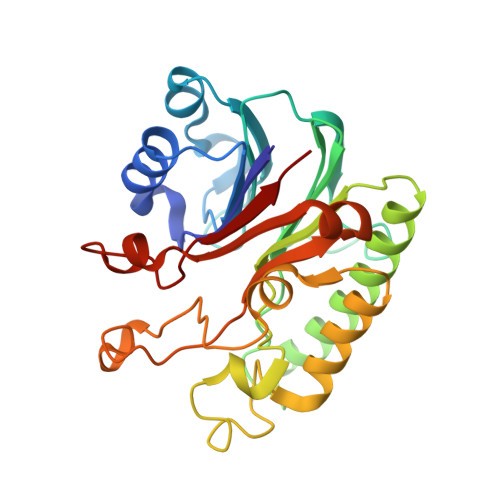
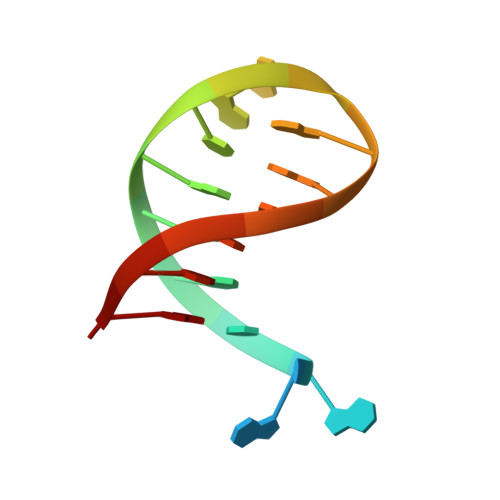
Structural basis for recognition and repair of the 3'-phosphate by NExo, a base excision DNA repair nuclease from Neisseria meningitidis.
Silhan, J., Zhao, Q., Boura, E., Thomson, H., Forster, A., Tang, C.M., Freemont, P.S., Baldwin, G.S.(2018) Nucleic Acids Res 46: 11980-11989
- PubMed: 30329088
- DOI: https://doi.org/10.1093/nar/gky934
- Primary Citation of Related Structures:
6FK4, 6FK5, 6FKE - PubMed Abstract:
NExo is an enzyme from Neisseria meningitidis that is specialized in the removal of the 3'-phosphate and other 3'-lesions, which are potential blocks for DNA repair. NExo is a highly active DNA 3'-phosphatase, and although it is from the class II AP family it lacks AP endonuclease activity. In contrast, the NExo homologue NApe, lacks 3'-phosphatase activity but is an efficient AP endonuclease. These enzymes act together to protect the meningococcus from DNA damage arising mainly from oxidative stress and spontaneous base loss. In this work, we present crystal structures of the specialized 3'-phosphatase NExo bound to DNA in the presence and absence of a 3'-phosphate lesion. We have outlined the reaction mechanism of NExo, and using point mutations we bring mechanistic insights into the specificity of the 3'-phosphatase activity of NExo. Our data provide further insight into the molecular origins of plasticity in substrate recognition for this class of enzymes. From this we hypothesize that these specialized enzymes lead to enhanced efficiency and accuracy of DNA repair and that this is important for the biological niche occupied by this bacterium.
- Institute of Organic Chemistry and Biochemistry of the Czech Academy of Sciences, Czech Republic.
Organizational Affiliation: