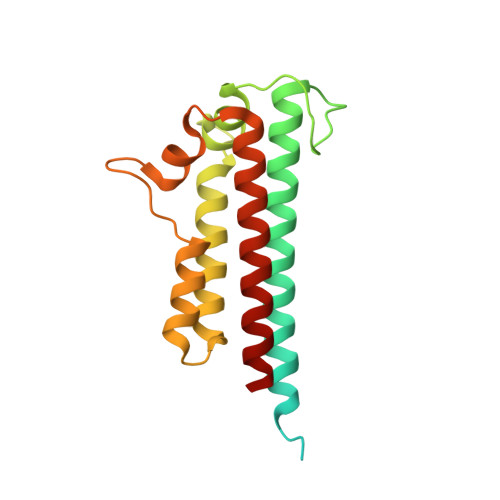
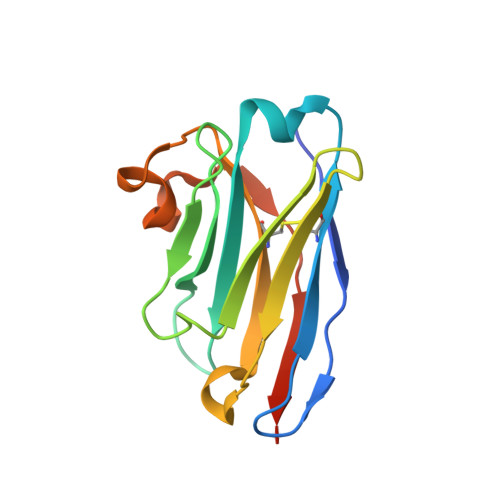
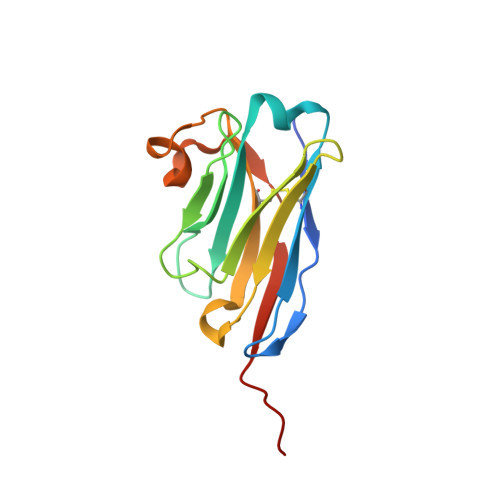
Type IX secretion system PorM and gliding machinery GldM form arches spanning the periplasmic space.
Leone, P., Roche, J., Vincent, M.S., Tran, Q.H., Desmyter, A., Cascales, E., Kellenberger, C., Cambillau, C., Roussel, A.(2018) Nat Commun 9: 429-429
- PubMed: 29382829
- DOI: https://doi.org/10.1038/s41467-017-02784-7
- Primary Citation of Related Structures:
6EY0, 6EY4, 6EY5, 6EY6 - PubMed Abstract:
Type IX secretion system (T9SS), exclusively present in the Bacteroidetes phylum, has been studied mainly in Flavobacterium johnsoniae and Porphyromonas gingivalis. Among the 18 genes, essential for T9SS function, a group of four, porK-N (P. gingivalis) or gldK-N (F. johnsoniae) belongs to a co-transcribed operon that expresses the T9SS core membrane complex. The central component of this complex, PorM (or GldM), is anchored in the inner membrane by a trans-membrane helix and interacts through the outer membrane PorK-N complex. There is a complete lack of available atomic structures for any component of T9SS, including the PorKLMN complex. Here we report the crystal structure of the GldM and PorM periplasmic domains. Dimeric GldM and PorM, each contain four domains of ~180-Å length that span most of the periplasmic space. These and previously reported results allow us to propose a model of the T9SS core membrane complex as well as its functional behavior.
- Architecture et Fonction des Macromolécules Biologiques, Aix-Marseille Université, UMR 7257, 163 Avenue de Luminy, Case 932, 13009, Marseille, France.
Organizational Affiliation: