Crystal structures of thrombin in complex with chemically modified thrombin DNA aptamers reveal the origins of enhanced affinity.
Dolot, R., Lam, C.H., Sierant, M., Zhao, Q., Liu, F.W., Nawrot, B., Egli, M., Yang, X.(2018) Nucleic Acids Res 46: 4819-4830
- PubMed: 29684204
- DOI: https://doi.org/10.1093/nar/gky268
- Primary Citation of Related Structures:
6EO6, 6EO7 - PubMed Abstract:
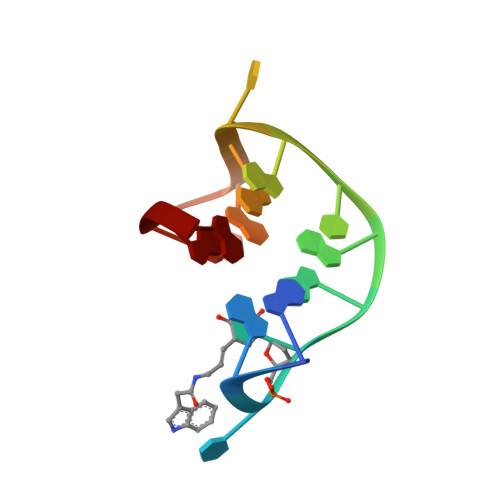
Thrombin-binding aptamer (TBA) is a DNA 15-mer of sequence 5'-GGT TGG TGT GGT TGG-3' that folds into a G-quadruplex structure linked by two T-T loops located on one side and a T-G-T loop on the other. These loops are critical for post-SELEX modification to improve TBA target affinity. With this goal in mind we synthesized a T analog, 5-(indolyl-3-acetyl-3-amino-1-propenyl)-2'-deoxyuridine (W) to substitute one T or a pair of Ts. Subsequently, the affinity for each analog was determined by biolayer interferometry. An aptamer with W at position 4 exhibited about 3-fold increased binding affinity, and replacing both T4 and T12 with W afforded an almost 10-fold enhancement compared to native TBA. To better understand the role of the substituent's aromatic moiety, an aptamer with 5-(methyl-3-acetyl-3-amino-1-propenyl)-2'-deoxyuridine (K; W without the indole moiety) in place of T4 was also synthesized. This K4 aptamer was found to improve affinity 7-fold relative to native TBA. Crystal structures of aptamers with T4 replaced by either W or K bound to thrombin provide insight into the origins of the increased affinities. Our work demonstrates that facile chemical modification of a simple DNA aptamer can be used to significantly improve its binding affinity for a well-established pharmacological target protein.
- Centre of Molecular and Macromolecular Studies, Polish Academy of Sciences, 90-363 Lodz, Sienkiewicza 112, Poland.
Organizational Affiliation: