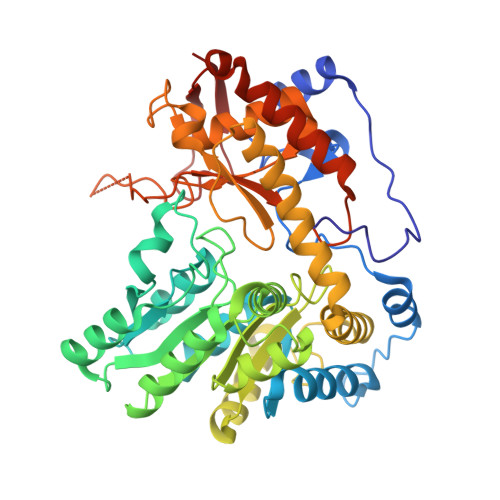
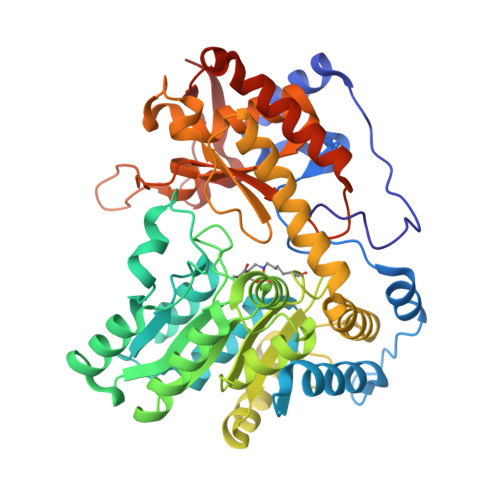
Crystal Structures of Wild-Type and F448A Mutant Citrobacter freundii Tyrosine Phenol-Lyase Complexed with a Substrate and Inhibitors: Implications for the Reaction Mechanism.
Phillips, R.S., Craig, S.(2018) Biochemistry 57: 6166-6179
- PubMed: 30260636
- DOI: https://doi.org/10.1021/acs.biochem.8b00724
- Primary Citation of Related Structures:
6DUR, 6DVX, 6DXV, 6DYT, 6DZ5, 6ECG - PubMed Abstract:
Tyrosine phenol-lyase (TPL; EC 4.1.99.2) is a pyridoxal 5'-phosphate-dependent enzyme that catalyzes the reversible hydrolytic cleavage of l-tyrosine to phenol and ammonium pyruvate. We have shown previously that F448A TPL has k cat and k cat / K m values for l-tyrosine reduced by ∼10 4 -fold [Phillips, R. S., Vita, A., Spivey, J. B., Rudloff, A. P., Driscoll, M. D., and Hay, S. (2016) ACS Catal. 6, 6770-6779]. We have now obtained crystal structures of F448A TPL and complexes with l-alanine, l-methionine, l-phenylalanine, and 3-F-l-tyrosine at 2.05-2.27 Å and the complex of wild-type TPL with l-phenylalanine at 1.8 Å. The small domain of F448A TPL, where Phe-448 is located, is more disordered in chain A than in wild-type TPL. The complexes of F448A TPL with l-alanine and l-phenylalanine are in an open conformation in both chains, while the complex with l-methionine is a 52:48 open:closed equilibrium mixture in chain A. Wild-type TPL with l-alanine is closed in chain A and open in chain B, and the complex with l-phenylalanine is a 56:44 open:closed mixture in chain A. Thus, the Phe-448 to alanine mutation affects the conformational equilibrium of open and closed active sites. The structure of the 3-F-l-tyrosine quinonoid complex of F448A TPL is unstrained and in an open conformation, with a hydrogen bond from the phenolic OH to Thr-124. These results support our previous conclusion that ground-state strain plays a critical role in the mechanism of TPL.