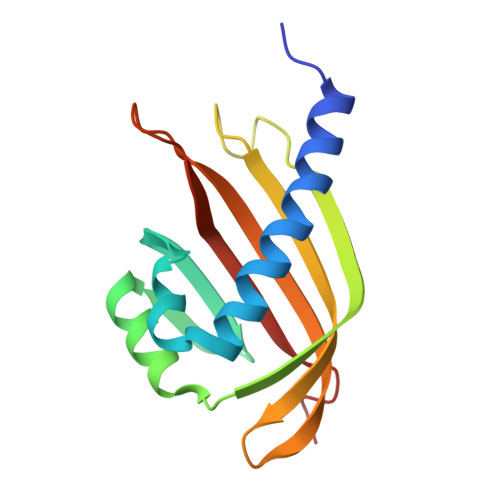
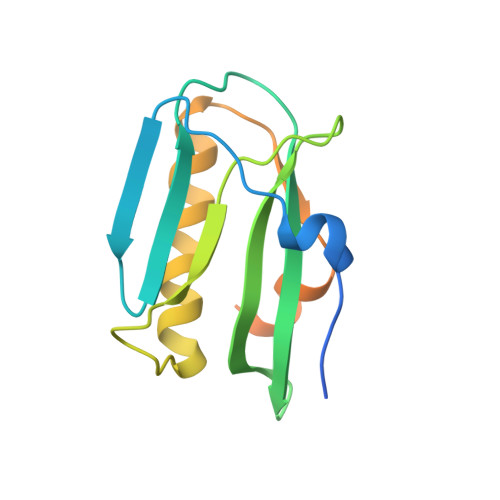
Structural basis for influenza virus NS1 protein block of mRNA nuclear export.
Zhang, K., Xie, Y., Munoz-Moreno, R., Wang, J., Zhang, L., Esparza, M., Garcia-Sastre, A., Fontoura, B.M.A., Ren, Y.(2019) Nat Microbiol 4: 1671-1679
- PubMed: 31263181
- DOI: https://doi.org/10.1038/s41564-019-0482-x
- Primary Citation of Related Structures:
6E5U - PubMed Abstract:
Influenza viruses antagonize key immune defence mechanisms via the virulence factor non-structural protein 1 (NS1). A key mechanism of virulence by NS1 is blocking nuclear export of host messenger RNAs, including those encoding immune factors 1-3 ; however, the direct cellular target of NS1 and the mechanism of host mRNA export inhibition are not known. Here, we identify the target of NS1 as the mRNA export receptor complex, nuclear RNA export factor 1-nuclear transport factor 2-related export protein 1 (NXF1-NXT1), which is the principal receptor mediating docking and translocation of mRNAs through the nuclear pore complex via interactions with nucleoporins 4,5 . We determined the crystal structure of NS1 in complex with NXF1-NXT1 at 3.8 Å resolution. The structure reveals that NS1 prevents binding of NXF1-NXT1 to nucleoporins, thereby inhibiting mRNA export through the nuclear pore complex into the cytoplasm for translation. We demonstrate that a mutant influenza virus deficient in binding NXF1-NXT1 does not block host mRNA export and is attenuated. This attenuation is marked by the release of mRNAs encoding immune factors from the nucleus. In sum, our study uncovers the molecular basis of a major nuclear function of influenza NS1 protein that causes potent blockage of host gene expression and contributes to inhibition of host immunity.
- Department of Cell Biology, University of Texas Southwestern Medical Center, Dallas, TX, USA.
Organizational Affiliation: