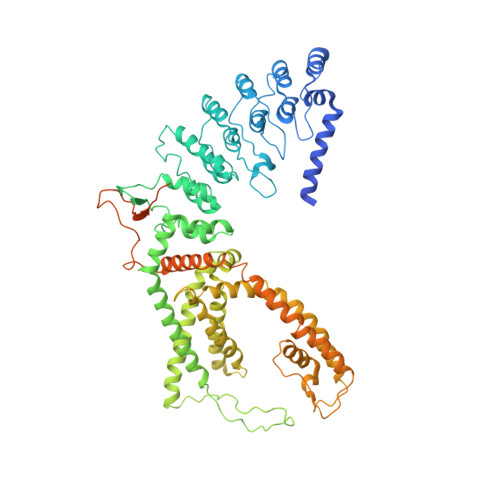
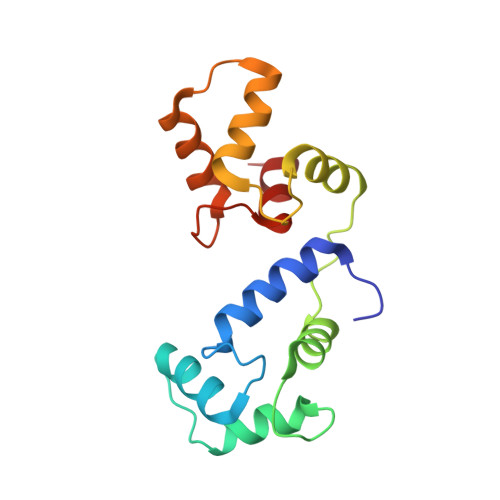
Mechanism of calmodulin inactivation of the calcium-selective TRP channel TRPV6.
Singh, A.K., McGoldrick, L.L., Twomey, E.C., Sobolevsky, A.I.(2018) Sci Adv 4: eaau6088-eaau6088
- PubMed: 30116787
- DOI: https://doi.org/10.1126/sciadv.aau6088
- Primary Citation of Related Structures:
6E2F, 6E2G - PubMed Abstract:
Calcium (Ca 2+ ) plays a major role in numerous physiological processes. Ca 2+ homeostasis is tightly controlled by ion channels, the aberrant regulation of which results in various diseases including cancers. Calmodulin (CaM)-mediated Ca 2+ -induced inactivation is an ion channel regulatory mechanism that protects cells against the toxic effects of Ca 2+ overload. We used cryo-electron microscopy to capture the epithelial calcium channel TRPV6 (transient receptor potential vanilloid subfamily member 6) inactivated by CaM. The TRPV6-CaM complex exhibits 1:1 stoichiometry; one TRPV6 tetramer binds both CaM lobes, which adopt a distinct head-to-tail arrangement. The CaM carboxyl-terminal lobe plugs the channel through a unique cation-π interaction by inserting the side chain of lysine K115 into a tetra-tryptophan cage at the pore's intracellular entrance. We propose a mechanism of CaM-mediated Ca 2+ -induced inactivation that can be explored for therapeutic design.
- Department of Biochemistry and Molecular Biophysics, Columbia University, 650 West 168th Street, New York, NY 10032, USA.
Organizational Affiliation: