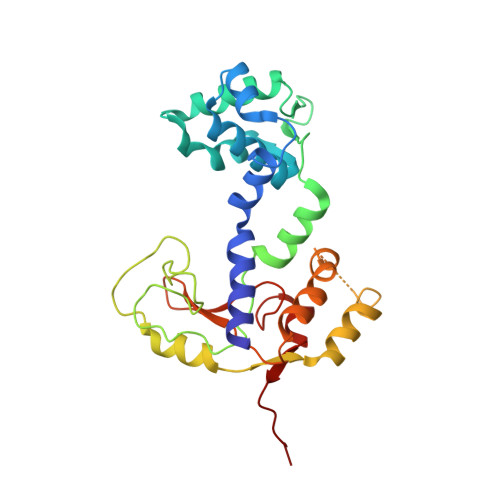
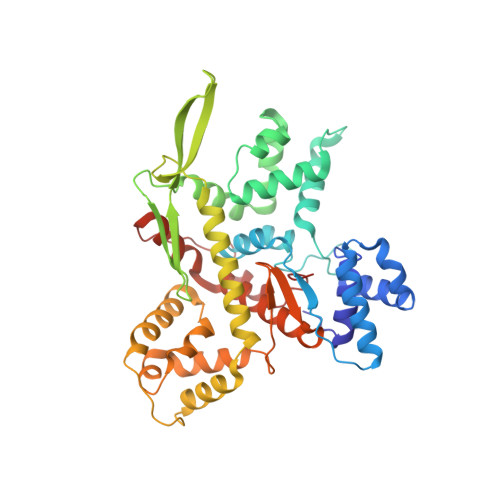
Minimalism and functionality: Structural lessons from the heterodimeric N4 bacteriophage RNA polymerase II.
Molodtsov, V., Murakami, K.S.(2018) J Biological Chem 293: 13616-13625
- PubMed: 29991593
- DOI: https://doi.org/10.1074/jbc.RA118.003447
- Primary Citation of Related Structures:
6DT7, 6DT8, 6DTA - PubMed Abstract:
Genomes of phages, mitochondria, and chloroplasts are transcribed by a diverse group of transcriptional machineries with structurally related single-subunit RNA polymerases (RNAPs). Our understanding of transcription mechanisms of these enzymes is predominantly based on biochemical and structural studies of three most-studied members, transcription factor-independent phage T7 RNAP, transcription factor-dependent phage N4 virion-encapsidated RNAP, and transcription factor-dependent mitochondrial RNAPs (mtRNAP). Although these RNAPs employ completely different mechanisms for promoter recognition and transcription termination, these enzymes are relatively large and formed by single polypeptides. Historically being a model enzyme for studying the mechanisms of transcription by T7-like RNAPs, however, T7 RNAP represents only a small group of RNAPs in this family. The vast majority of T7-like RNAPs are transcription factor-dependent, and several of them are heterodimeric enzymes. Here, we report X-ray crystal structures of transcription complexes of the smallest and heterodimeric form of T7-like RNAP, bacteriophage N4 RNAPII, providing insights into the structural organization of a minimum RNAP in this family. We analyze structural and functional aspects of heterodimeric architecture of N4 RNAPII concerning the mechanisms of transcription initiation and transition to processive RNA elongation. Interestingly, N4 RNAPII maintains the same conformation in promoter-bound and elongation transcription complexes, revealing a novel transcription mechanism for single-subunit RNAPs. This work establishes a structural basis for studying mechanistic aspects of transcription by factor-dependent minimum RNAP.
- From the Department of Biochemistry and Molecular Biology, The Center for RNA Molecular Biology, The Pennsylvania State University, University Park, Pennsylvania 16802 vum5@psu.edu.
Organizational Affiliation: