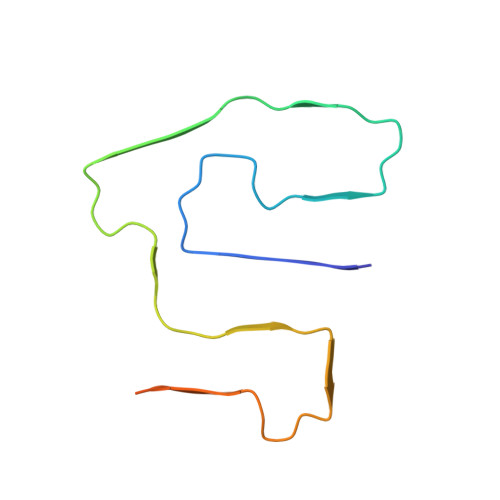
Cryo-EM fibril structures from systemic AA amyloidosis reveal the species complementarity of pathological amyloids.
Liberta, F., Loerch, S., Rennegarbe, M., Schierhorn, A., Westermark, P., Westermark, G.T., Hazenberg, B.P.C., Grigorieff, N., Fandrich, M., Schmidt, M.(2019) Nat Commun 10: 1104-1104
- PubMed: 30846696
- DOI: https://doi.org/10.1038/s41467-019-09033-z
- Primary Citation of Related Structures:
6DSO, 6MST - PubMed Abstract:
Systemic AA amyloidosis is a worldwide occurring protein misfolding disease of humans and animals. It arises from the formation of amyloid fibrils from the acute phase protein serum amyloid A. Here, we report the purification and electron cryo-microscopy analysis of amyloid fibrils from a mouse and a human patient with systemic AA amyloidosis. The obtained resolutions are 3.0 Å and 2.7 Å for the murine and human fibril, respectively. The two fibrils differ in fundamental properties, such as presence of right-hand or left-hand twisted cross-β sheets and overall fold of the fibril proteins. Yet, both proteins adopt highly similar β-arch conformations within the N-terminal ~21 residues. Our data demonstrate the importance of the fibril protein N-terminus for the stability of the analyzed amyloid fibril morphologies and suggest strategies of combating this disease by interfering with specific fibril polymorphs.
- Institute of Protein Biochemistry, Ulm University, 89081, Ulm, Germany.
Organizational Affiliation: