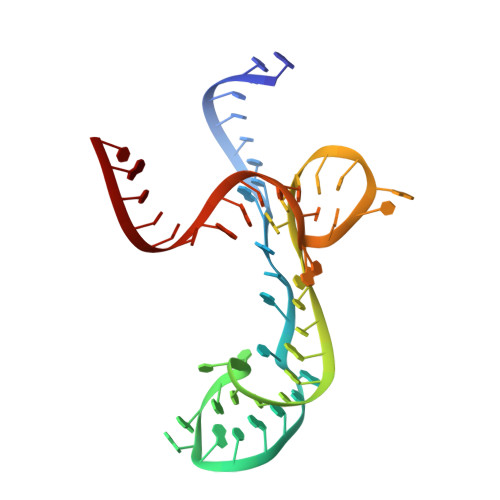
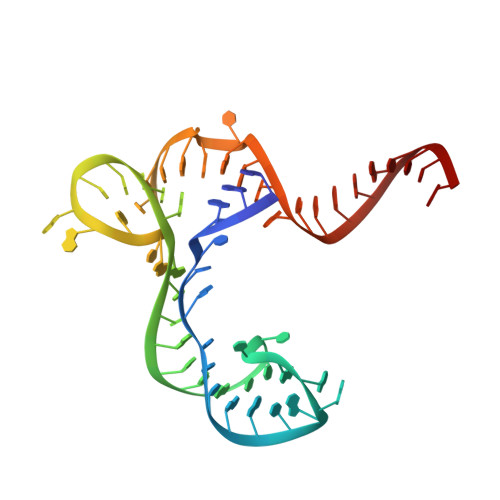
Structure-Activity Relationship of Flavin Analogues That Target the Flavin Mononucleotide Riboswitch.
Vicens, Q., Mondragon, E., Reyes, F.E., Coish, P., Aristoff, P., Berman, J., Kaur, H., Kells, K.W., Wickens, P., Wilson, J., Gadwood, R.C., Schostarez, H.J., Suto, R.K., Blount, K.F., Batey, R.T.(2018) ACS Chem Biol 13: 2908-2919
- PubMed: 30107111
- DOI: https://doi.org/10.1021/acschembio.8b00533
- Primary Citation of Related Structures:
6DN1, 6DN2, 6DN3 - PubMed Abstract:
The flavin mononucleotide (FMN) riboswitch is an emerging target for the development of novel RNA-targeting antibiotics. We previously discovered an FMN derivative, 5FDQD, that protects mice against diarrhea-causing Clostridium difficile bacteria. Here, we present the structure-based drug design strategy that led to the discovery of this fluoro-phenyl derivative with antibacterial properties. This approach involved the following stages: (1) structural analysis of all available free and bound FMN riboswitch structures; (2) design, synthesis, and purification of derivatives; (3) in vitro testing for productive binding using two chemical probing methods; (4) in vitro transcription termination assays; and (5) resolution of the crystal structures of the FMN riboswitch in complex with the most mature candidates. In the process, we delineated principles for productive binding to this riboswitch, thereby demonstrating the effectiveness of a coordinated structure-guided approach to designing drugs against RNA.
- Department of Chemistry and Biochemistry , University of Colorado , 596 UCB , Boulder , Colorado 80309 , United States.
Organizational Affiliation: