Structure of Eremothecium gossypii Shu1:Shu2 complex
Singh, N., Corbett, K.D.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Suppressor of hydroxyurea sensitivity protein 1 | 148 | Eremothecium gossypii ATCC 10895 | Mutation(s): 0 Gene Names: AGOS_ABR011W | 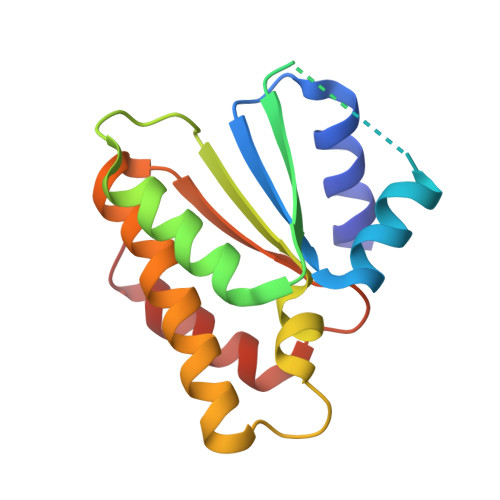 | |
UniProt | |||||
Find proteins for Q75DL0 (Eremothecium gossypii (strain ATCC 10895 / CBS 109.51 / FGSC 9923 / NRRL Y-1056)) Explore Q75DL0 Go to UniProtKB: Q75DL0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q75DL0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Suppressor of hydroxyurea sensitivity protein 2 | 201 | Eremothecium gossypii ATCC 10895 | Mutation(s): 0 Gene Names: SHU2, AGR140C | 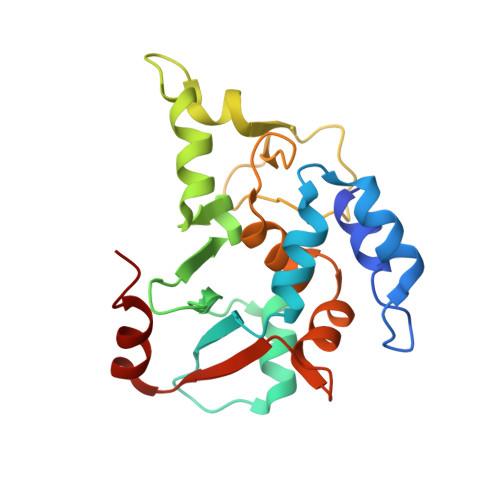 | |
UniProt | |||||
Find proteins for Q74ZQ8 (Eremothecium gossypii (strain ATCC 10895 / CBS 109.51 / FGSC 9923 / NRRL Y-1056)) Explore Q74ZQ8 Go to UniProtKB: Q74ZQ8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q74ZQ8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NHE Query on NHE | D [auth A] | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID C8 H17 N O3 S MKWKNSIESPFAQN-UHFFFAOYSA-N |  | ||
| ZN Query on ZN | E [auth B] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| MG Query on MG | C [auth A] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 37.252 | α = 90 |
| b = 83.909 | β = 90 |
| c = 107.082 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| Aimless | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | R01 GM104141 |