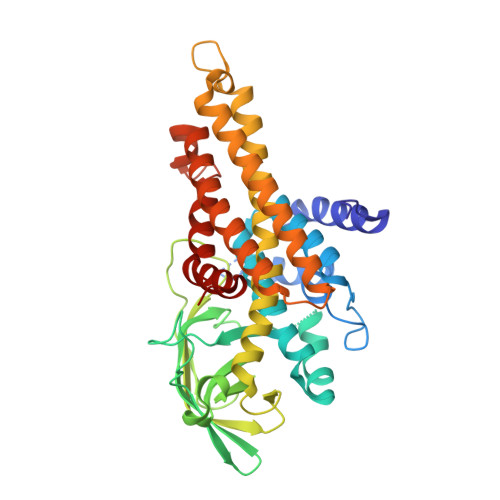
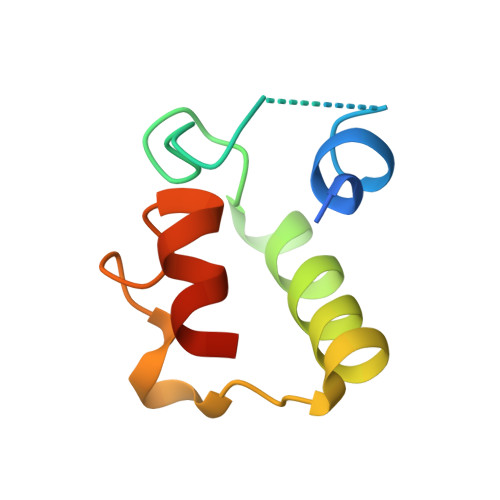
Insights into Thiotemplated Pyrrole Biosynthesis Gained from the Crystal Structure of Flavin-Dependent Oxidase in Complex with Carrier Protein.
Thapa, H.R., Robbins, J.M., Moore, B.S., Agarwal, V.(2019) Biochemistry 58: 918-929
- PubMed: 30620182
- DOI: https://doi.org/10.1021/acs.biochem.8b01177
- Primary Citation of Related Structures:
6CXT, 6CY8 - PubMed Abstract:
Sequential enzymatic reactions on substrates tethered to carrier proteins (CPs) generate thiotemplated building blocks that are then delivered to nonribosomal peptide synthetases (NRPSs) to generate peptidic natural products. The underlying diversity of these thiotemplated building blocks is the principal driver of the chemical diversity of NRPS-derived natural products. Structural insights into recognition of CPs by tailoring enzymes that generate these building blocks are sparse. Here we present the crystal structure of a flavin-dependent prolyl oxidase that furnishes thiotemplated pyrrole as the product, in complex with its cognate CP in the holo and product-bound states. The thiotemplated pyrrole is an intermediate that is well-represented in natural product biosynthetic pathways. Our results delineate the interactions between the CP and the oxidase while also providing insights into the stereospecificity of the enzymatic oxidation of the prolyl heterocycle to the aromatic pyrrole. Biochemical validation of the interaction between the CP and the oxidase demonstrates that NRPSs recognize and bind to their CPs using interactions quite different from those of fatty acid and polyketide biosynthetic enzymes. Our results posit that structural diversity in natural product biosynthesis can be, and is, derived from subtle modifications of primary metabolic enzymes.
- School of Chemistry and Biochemistry , Georgia Institute of Technology , Atlanta , Georgia 30332 , United States.
Organizational Affiliation: