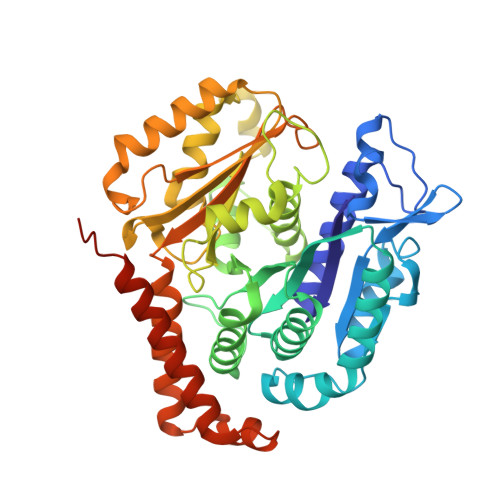
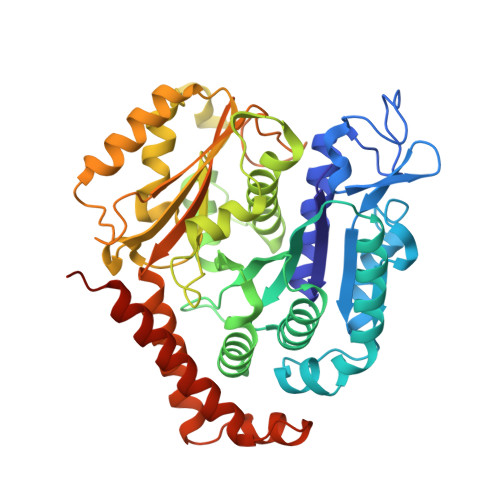
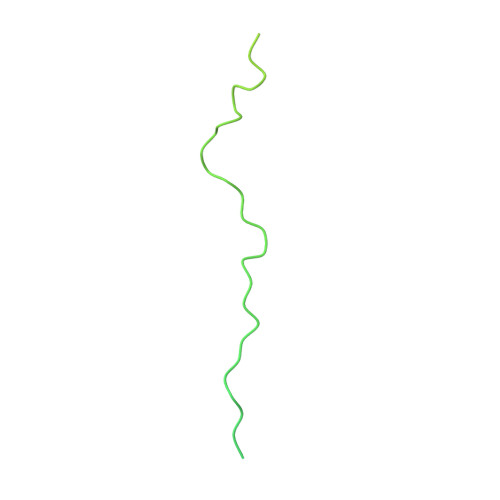
Near-atomic model of microtubule-tau interactions.
Kellogg, E.H., Hejab, N.M.A., Poepsel, S., Downing, K.H., DiMaio, F., Nogales, E.(2018) Science 360: 1242-1246
- PubMed: 29748322
- DOI: https://doi.org/10.1126/science.aat1780
- Primary Citation of Related Structures:
6CVJ, 6CVN - PubMed Abstract:
Tau is a developmentally regulated axonal protein that stabilizes and bundles microtubules (MTs). Its hyperphosphorylation is thought to cause detachment from MTs and subsequent aggregation into fibrils implicated in Alzheimer's disease. It is unclear which tau residues are crucial for tau-MT interactions, where tau binds on MTs, and how it stabilizes them. We used cryo-electron microscopy to visualize different tau constructs on MTs and computational approaches to generate atomic models of tau-tubulin interactions. The conserved tubulin-binding repeats within tau adopt similar extended structures along the crest of the protofilament, stabilizing the interface between tubulin dimers. Our structures explain the effect of phosphorylation on MT affinity and lead to a model of tau repeats binding in tandem along protofilaments, tethering together tubulin dimers and stabilizing polymerization interfaces.
- QB3 Institute and Department of Molecular and Cell Biology, University of California-Berkeley, Berkeley, CA 94720, USA.
Organizational Affiliation: