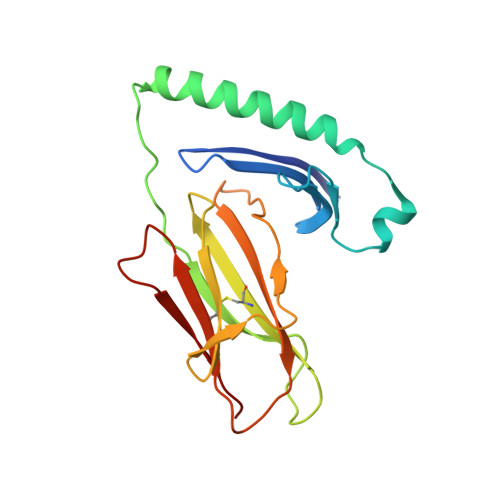
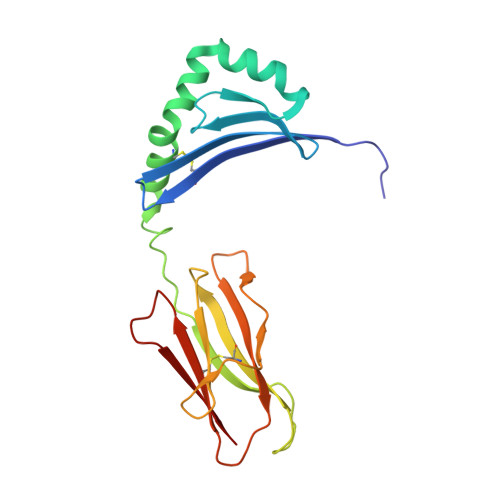
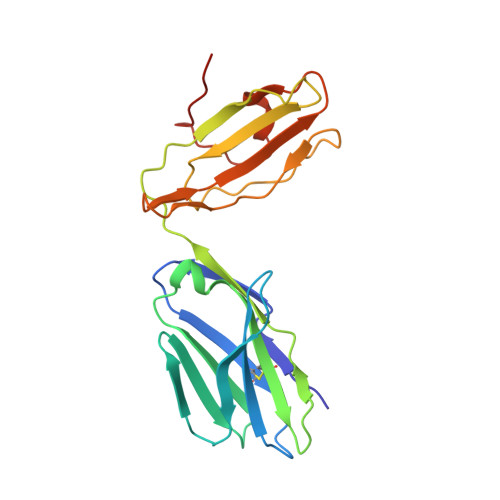
CD4+T cell-mediated HLA class II cross-restriction in HIV controllers.
Galperin, M., Farenc, C., Mukhopadhyay, M., Jayasinghe, D., Decroos, A., Benati, D., Tan, L.L., Ciacchi, L., Reid, H.H., Rossjohn, J., Chakrabarti, L.A., Gras, S.(2018) Sci Immunol 3
- PubMed: 29884618
- DOI: https://doi.org/10.1126/sciimmunol.aat0687
- Primary Citation of Related Structures:
6CPH, 6CPL, 6CPN, 6CPO, 6CQJ, 6CQL, 6CQN, 6CQQ, 6CQR - PubMed Abstract:
Rare individuals, termed HIV controllers, spontaneously control HIV infection by mounting efficient T cell responses against the virus. Protective CD4 + T cell responses from HIV controllers involve high-affinity public T cell receptors (TCRs) recognizing an immunodominant capsid epitope (Gag293) presented by a remarkably broad array of human leukocyte antigen (HLA) class II molecules. Here, we determine the structures of a prototypical public TCR bound to HLA-DR1, HLA-DR11, and HLA-DR15 molecules presenting the Gag293 epitope. TCR recognition was driven by contacts with the Gag293 epitope, a feature that underpinned the extensive HLA cross-restriction. These high-affinity TCRs promoted mature immunological synapse formation and cytotoxic capacity in both CD4 + and CD8 + T cells. The public TCRs suppressed HIV replication in multiple genetic backgrounds ex vivo, emphasizing the functional advantage conferred by broad HLA class II cross-restriction.
- Pasteur Institute, Viral Pathogenesis Unit, Paris, France.
Organizational Affiliation: