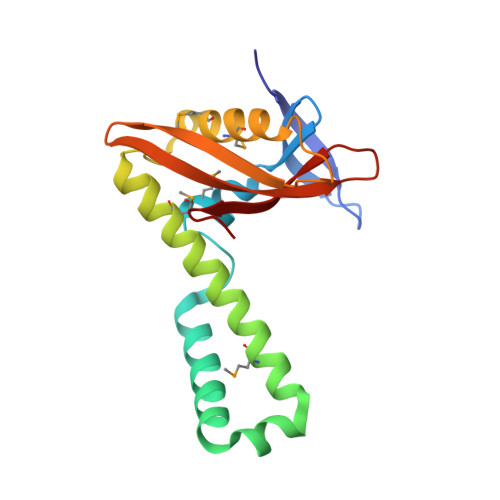
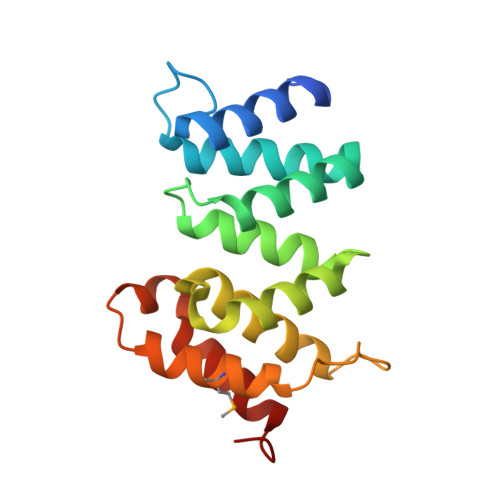
Convergent Evolution of the Barnase/EndoU/Colicin/RelE (BECR) Fold in Antibacterial tRNase Toxins.
Gucinski, G.C., Michalska, K., Garza-Sanchez, F., Eschenfeldt, W.H., Stols, L., Nguyen, J.Y., Goulding, C.W., Joachimiak, A., Hayes, C.S.(2019) Structure 27: 1660
- PubMed: 31515004
- DOI: https://doi.org/10.1016/j.str.2019.08.010
- Primary Citation of Related Structures:
6CP8, 6CP9 - PubMed Abstract:
Contact-dependent growth inhibition (CDI) is a form of interbacterial competition mediated by CdiB-CdiA two-partner secretion systems. CdiA effector proteins carry polymorphic C-terminal toxin domains (CdiA-CT), which are neutralized by specific CdiI immunity proteins to prevent self-inhibition. Here, we present the crystal structures of CdiA-CT⋅CdiI complexes from Klebsiella pneumoniae 342 and Escherichia coli 3006. The toxins adopt related folds that resemble the ribonuclease domain of colicin D, and both are isoacceptor-specific tRNases that cleave the acceptor stem of deacylated tRNA GAU Ile . Although the toxins are similar in structure and substrate specificity, CdiA-CT Kp342 activity requires translation factors EF-Tu and EF-Ts, whereas CdiA-CT EC3006 is intrinsically active. Furthermore, the corresponding immunity proteins are unrelated in sequence and structure. CdiI Kp342 forms a dimeric β sandwich, whereas CdiI EC3006 is an α-solenoid monomer. Given that toxin-immunity genes co-evolve as linked pairs, these observations suggest that the similarities in toxin structure and activity reflect functional convergence.
- Biomolecular Science and Engineering Program, University of California, Santa Barbara, CA, USA.
Organizational Affiliation: