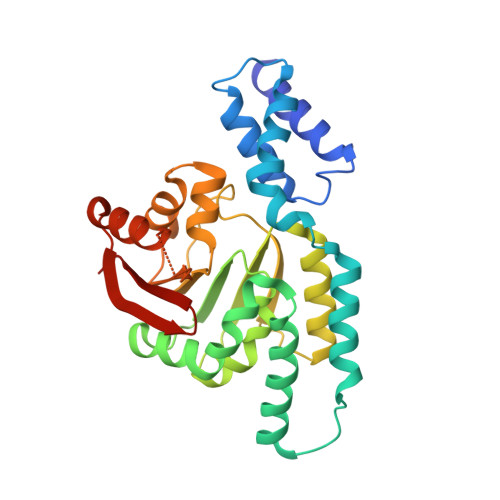
2.95 Angstrom Crystal Structure of 16S rRNA Methylase from Proteus mirabilis.
Minasov, G., Wawrzak, Z., Di Leo, R., Evdokimova, E., Savchenko, A., Satchell, K.J.F., Joachimiak, A., Center for Structural Genomics of Infectious Diseases (CSGID)To be published.