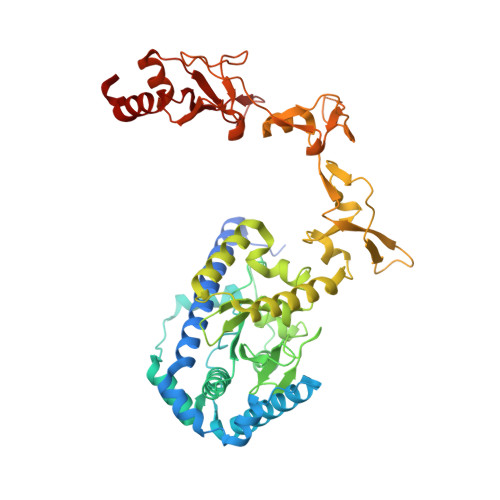
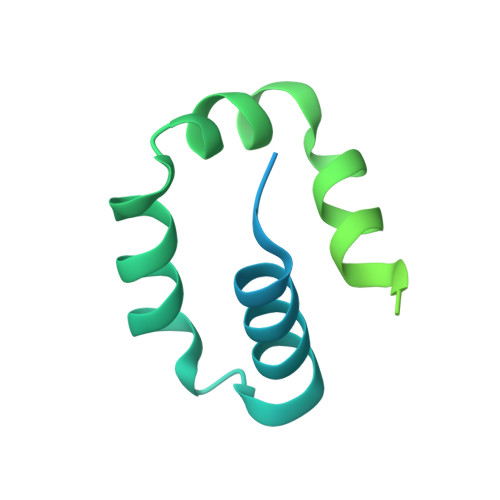
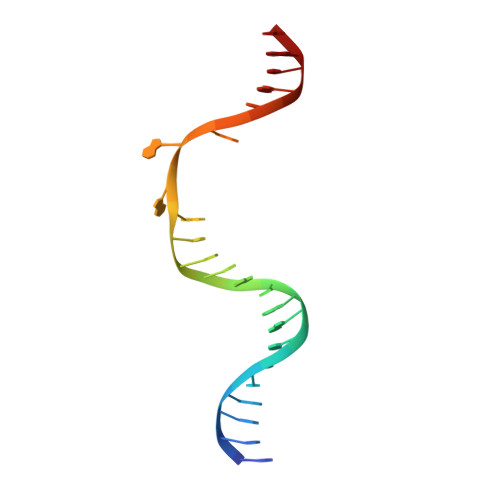
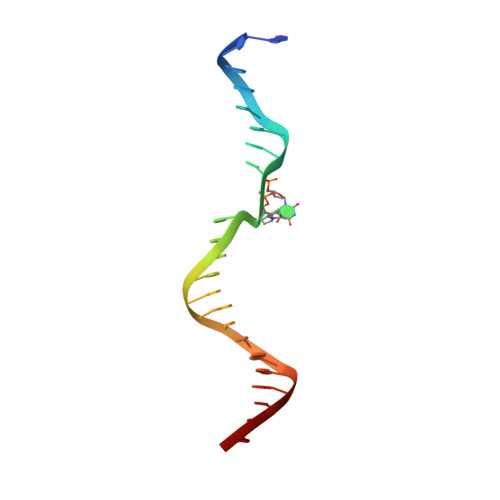
Structure and mechanism of pyrimidine-pyrimidone (6-4) photoproduct recognition by the Rad4/XPC nucleotide excision repair complex.
Paul, D., Mu, H., Zhao, H., Ouerfelli, O., Jeffrey, P.D., Broyde, S., Min, J.H.(2019) Nucleic Acids Res 47: 6015-6028
- PubMed: 31106376
- DOI: https://doi.org/10.1093/nar/gkz359
- Primary Citation of Related Structures:
6CFI - PubMed Abstract:
Failure in repairing ultraviolet radiation-induced DNA damage can lead to mutations and cancer. Among UV-lesions, the pyrimidine-pyrimidone (6-4) photoproduct (6-4PP) is removed from the genome much faster than the cyclobutane pyrimidine dimer (CPD), owing to the more efficient recognition of 6-4PP by XPC-RAD23B, a key initiator of global-genome nucleotide excision repair (NER). Here, we report a crystal structure of a Rad4-Rad23 (yeast XPC-Rad23B ortholog) bound to 6-4PP-containing DNA and 4-μs molecular dynamics (MD) simulations examining the initial binding of Rad4 to 6-4PP or CPD. This first structure of Rad4/XPC bound to a physiological substrate with matched DNA sequence shows that Rad4 flips out both 6-4PP-containing nucleotide pairs, forming an 'open' conformation. The MD trajectories detail how Rad4/XPC initiates 'opening' 6-4PP: Rad4 initially engages BHD2 to bend/untwist DNA from the minor groove, leading to unstacking and extrusion of the 6-4PP:AA nucleotide pairs towards the major groove. The 5' partner adenine first flips out and is captured by a BHD2/3 groove, while the 3' adenine extrudes episodically, facilitating ensuing insertion of the BHD3 β-hairpin to open DNA as in the crystal structure. However, CPD resists such Rad4-induced structural distortions. Untwisting/bending from the minor groove may be a common way to interrogate DNA in NER.
- Department of Chemistry & Biochemistry, Baylor University, Waco, TX 76798, USA.
Organizational Affiliation: