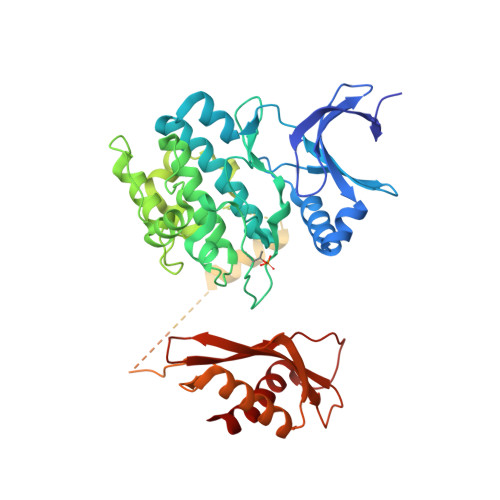
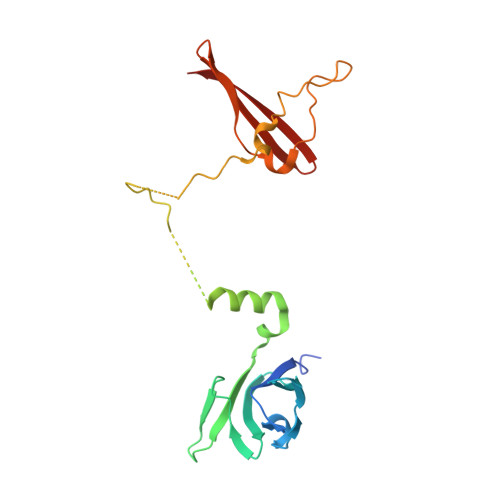
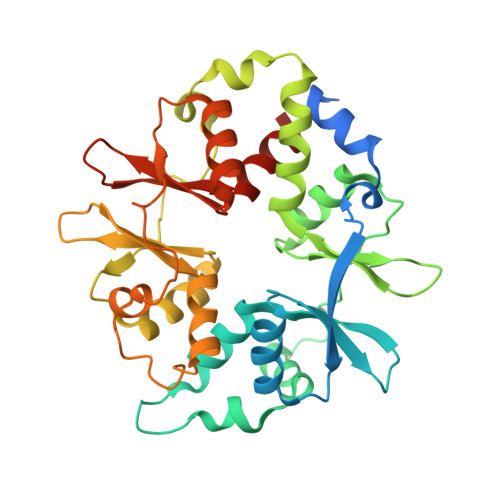
Structures of AMP-activated protein kinase bound to novel pharmacological activators in phosphorylated, non-phosphorylated, and nucleotide-free states.
Yan, Y., Zhou, X.E., Novick, S.J., Shaw, S.J., Li, Y., Brunzelle, J.S., Hitoshi, Y., Griffin, P.R., Xu, H.E., Melcher, K.(2019) J Biological Chem 294: 953-967
- PubMed: 30478170
- DOI: https://doi.org/10.1074/jbc.RA118.004883
- Primary Citation of Related Structures:
6C9F, 6C9G, 6C9H, 6C9J - PubMed Abstract:
AMP-activated protein kinase (AMPK) is an attractive therapeutic target for managing metabolic diseases. A class of pharmacological activators, including Merck 991, binds the AMPK ADaM site, which forms the interaction surface between the kinase domain (KD) of the α-subunit and the carbohydrate-binding module (CBM) of the β-subunit. Here, we report the development of two new 991-derivative compounds, R734 and R739, which potently activate AMPK in a variety of cell types, including β 2 -specific skeletal muscle cells. Surprisingly, we found that they have only minor effects on direct kinase activity of the recombinant α 1 β 2 γ 1 isoform yet robustly enhance protection against activation loop dephosphorylation. This mode of activation is reminiscent of that of ADP, which activates AMPK by binding to the nucleotide-binding sites in the γ-subunit, more than 60 Å away from the ADaM site. To understand the mechanisms of full and partial AMPK activation, we determined the crystal structures of fully active phosphorylated AMPK α 1 β 1 γ 1 bound to AMP and R734/R739 as well as partially active nonphosphorylated AMPK bound to R734 and AMP and phosphorylated AMPK bound to R734 in the absence of added nucleotides at <3-Å resolution. These structures and associated analyses identified a novel conformational state of the AMPK autoinhibitory domain associated with partial kinase activity and provide new insights into phosphorylation-dependent activation loop stabilization in AMPK.
- From the Center of Cancer and Cell Biology, Van Andel Research Institute, Grand Rapids, Michigan 49503.
Organizational Affiliation: