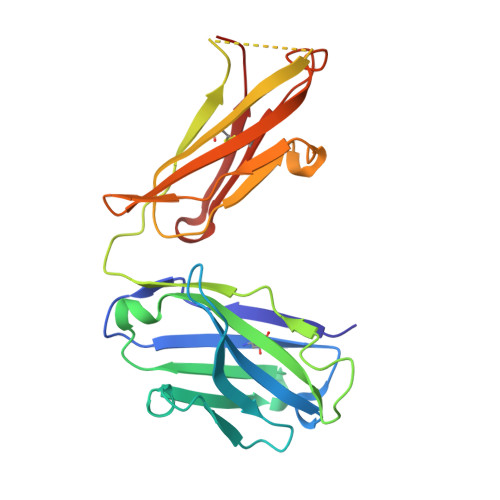
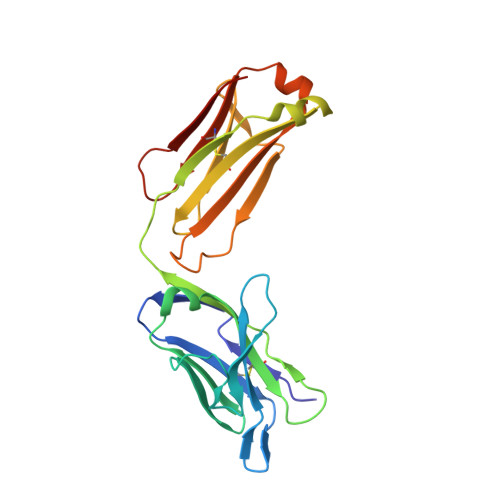
Immunogenetic and structural analysis of a class of HCV broadly neutralizing antibodies and their precursors.
Aleman, F., Tzarum, N., Kong, L., Nagy, K., Zhu, J., Wilson, I.A., Law, M.(2018) Proc Natl Acad Sci U S A 115: 7569-7574
- PubMed: 29954862
- DOI: https://doi.org/10.1073/pnas.1802378115
- Primary Citation of Related Structures:
6BZU, 6BZV, 6BZW, 6BZY - PubMed Abstract:
Elicitation of broadly neutralizing antibodies (bnAbs) is a leading strategy in rational vaccine design against antigenically diverse pathogens. Here, we studied a panel of monoclonal antibodies (mAbs) from mice immunized with the hepatitis C virus (HCV) envelope glycoproteins E1E2. Six of the mAbs recognize the conserved E2 antigenic site 412-423 (AS412) and cross-neutralize diverse HCV genotypes. Immunogenetic and structural analysis revealed that the antibodies originated from two different germline (GL) precursors and bind AS412 in a β-hairpin conformation. Intriguingly, the anti-HCV activity of one antibody lineage is associated with maturation of the light chain (LC), whereas the other lineage is dependent on heavy-chain (HC) maturation. Crystal structures of GL precursors of the LC-dependent lineage in complex with AS412 offer critical insights into the maturation process of bnAbs to HCV, providing a scientific foundation for utilizing the mouse model to study AS412-targeting vaccine candidates.
- Department of Immunology and Microbiology, The Scripps Research Institute, La Jolla, CA 92037.
Organizational Affiliation: