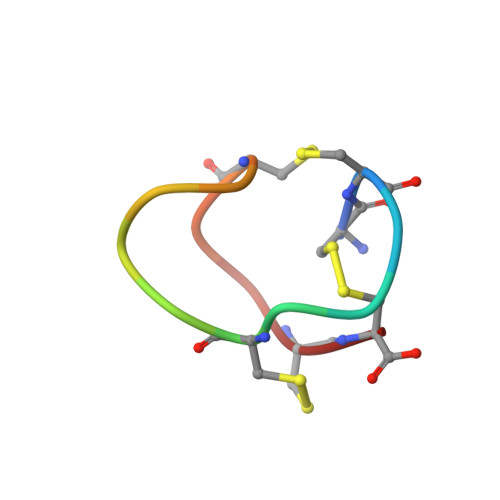
Structural plasticity of mini-M conotoxins - expression of all mini-M subtypes by Conus regius.
Franco, A., Dovell, S., Moller, C., Grandal, M., Clark, E., Mari, F.(2018) FEBS J 285: 887-902
- PubMed: 29283511
- DOI: https://doi.org/10.1111/febs.14372
- Primary Citation of Related Structures:
6BX9 - PubMed Abstract:
The mini-M conotoxins are peptidic scaffolds found in the venom of cones snails. These scaffolds are tightly folded structures held together by three disulfide bonds with a CC-C-C-CC arrangement (conotoxin framework III) and belong to the M Superfamily of conotoxins. Here, we describe mini-M conotoxins from the venom of Conus regius, a Western Atlantic worm-hunting cone snail species using transcriptomic and peptidomic analyses. These C. regius conotoxins belong to three different subtypes: M1, M2, and M3. The subtypes show little sequence homology, and their loop sizes (intercysteine amino acid chains) vary significantly. The mini-Ms isolated from dissected venom contains preferentially hydroxylated proline residues, thus augmenting the structural reach of this conotoxin class. Using 2D-NMR methods, we have determined the 3D structure of reg3b, an M2 subtype conotoxin, which shows a constrained multi-turn scaffold. The structural diversity found within mini-M conotoxin scaffolds of C. regius is indicative of structural hypervariability of the conotoxin M superfamily that is not seen in other superfamilies. These stable minimalistic scaffolds may be investigated for the development of engineered peptides for therapeutic applications. Sequences are available in GenBank under accession numbers MF588935-MF588952. Structural data are available in the RCSB protein database under the accession code 6BX9.
- Department of Chemistry and Biochemistry, Florida Atlantic University, Boca Raton, FL, USA.
Organizational Affiliation: