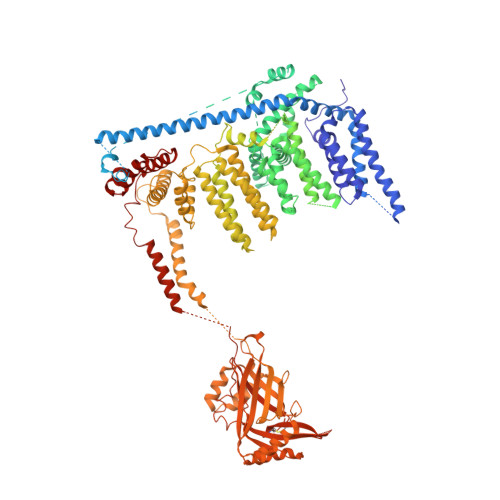
Structure of the mechanically activated ion channel Piezo1.
Saotome, K., Murthy, S.E., Kefauver, J.M., Whitwam, T., Patapoutian, A., Ward, A.B.(2018) Nature 554: 481-486
- PubMed: 29261642
- DOI: https://doi.org/10.1038/nature25453
- Primary Citation of Related Structures:
6BPZ - PubMed Abstract:
Piezo1 and Piezo2 are mechanically activated ion channels that mediate touch perception, proprioception and vascular development. Piezo proteins are distinct from other ion channels and their structure remains poorly defined, which impedes detailed study of their gating and ion permeation properties. Here we report a high-resolution cryo-electron microscopy structure of the mouse Piezo1 trimer. The detergent-solubilized complex adopts a three-bladed propeller shape with a curved transmembrane region containing at least 26 transmembrane helices per protomer. The flexible propeller blades can adopt distinct conformations, and consist of a series of four-transmembrane helical bundles that we term Piezo repeats. Carboxy-terminal domains line the central ion pore, and the channel is closed by constrictions in the cytosol. A kinked helical beam and anchor domain link the Piezo repeats to the pore, and are poised to control gating allosterically. The structure provides a foundation to dissect further how Piezo channels are regulated by mechanical force.
- Howard Hughes Medical Institute, Department of Neuroscience, The Scripps Research Institute, La Jolla, California 92037, USA.
Organizational Affiliation: