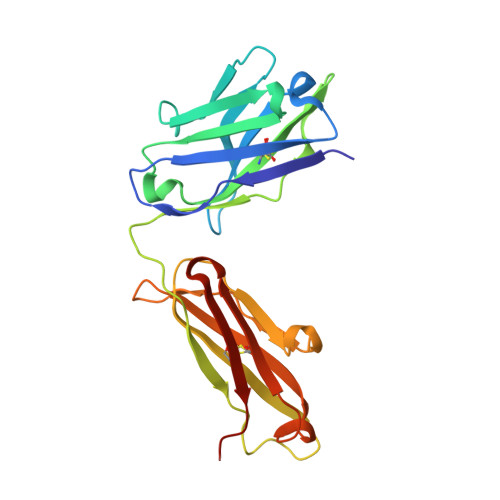
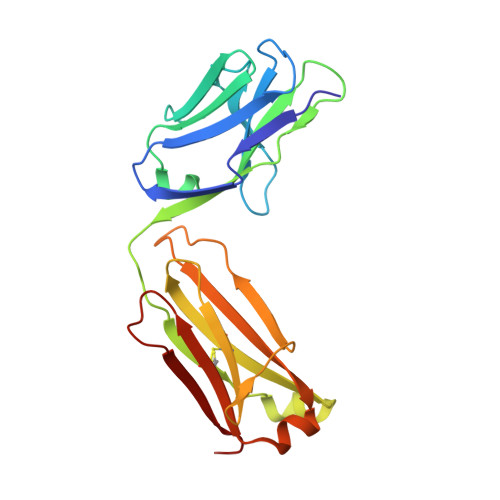
Tuning a Protein-Labeling Reaction to Achieve Highly Site Selective Lysine Conjugation.
Pham, G.H., Ou, W., Bursulaya, B., DiDonato, M., Herath, A., Jin, Y., Hao, X., Loren, J., Spraggon, G., Brock, A., Uno, T., Geierstanger, B.H., Cellitti, S.E.(2018) Chembiochem 19: 799-804
- PubMed: 29388367
- DOI: https://doi.org/10.1002/cbic.201700611
- Primary Citation of Related Structures:
6BHZ, 6BI0, 6BI2 - PubMed Abstract:
Activated esters are widely used to label proteins at lysine side chains and N termini. These reagents are useful for labeling virtually any protein, but robust reactivity toward primary amines generally precludes site-selective modification. In a unique case, fluorophenyl esters are shown to preferentially label human kappa antibodies at a single lysine (Lys188) within the light-chain constant domain. Neighboring residues His189 and Asp151 contribute to the accelerated rate of labeling at Lys188 relative to the ≈40 other lysine sites. Enriched Lys188 labeling can be enhanced from 50-70 % to >95 % by any of these approaches: lowering reaction temperature, applying flow chemistry, or mutagenesis of specific residues in the surrounding protein environment. Our results demonstrated that activated esters with fluoro-substituted aromatic leaving groups, including a fluoronaphthyl ester, can be generally useful reagents for site-selective lysine labeling of antibodies and other immunoglobulin-type proteins.
- Genomics Institute of the Novartis Research Foundation (GNF), Biotherapeutics & Biotechnology, 10675 John Jay Hopkins Drive, San Diego, CA, 92121, USA.
Organizational Affiliation: