Template-Catalyzed, Disulfide Conjugation of Monoclonal Antibodies Using a Natural Amino Acid Tag.
King, J.D., Ma, Y., Kuo, Y.C., Bzymek, K.P., Goodstein, L.H., Meyer, K., Moore, R.E., Crow, D., Colcher, D.M., Singh, G., Horne, D.A., Williams, J.C.(2018) Bioconjug Chem 29: 2074-2081
- PubMed: 29763554
- DOI: https://doi.org/10.1021/acs.bioconjchem.8b00284
- Primary Citation of Related Structures:
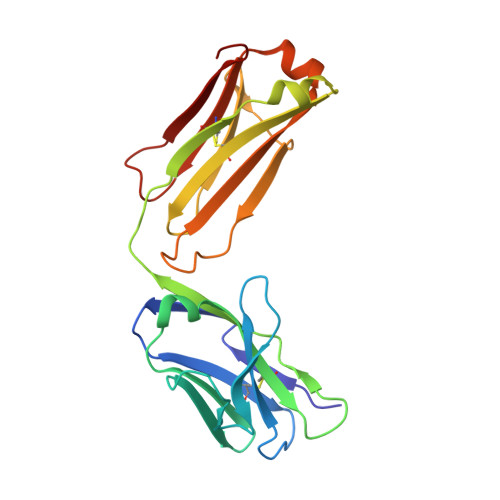
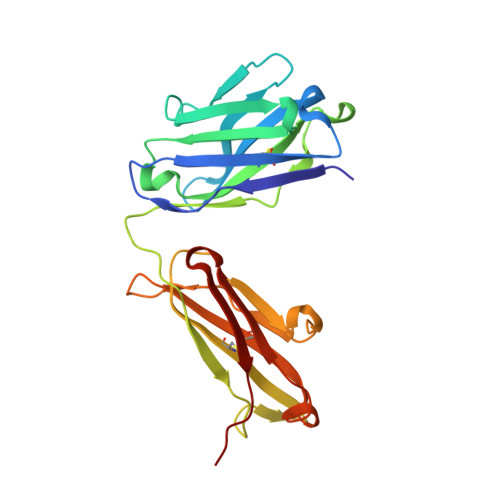
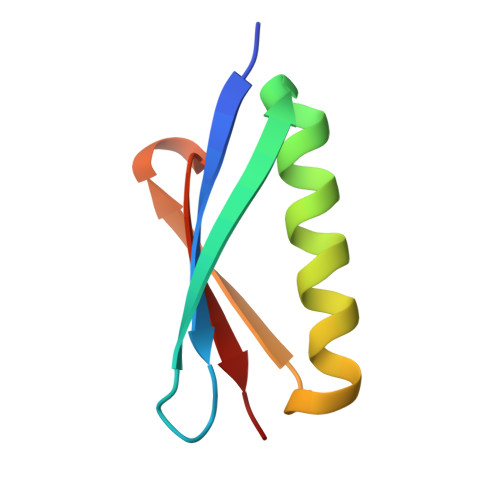
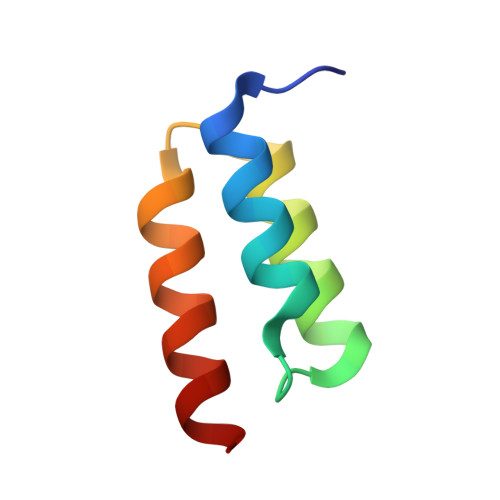
6B9Y, 6B9Z, 6BAE, 6BAH - PubMed Abstract:
The high specificity and favorable pharmacological properties of monoclonal antibodies (mAbs) have prompted significant interest in re-engineering this class of molecules to add novel functionalities for enhanced therapeutic and diagnostic potential. Here, we used the high affinity, meditope-Fab interaction to template and drive the rapid, efficient, and stable site-specific formation of a disulfide bond. We demonstrate that this template-catalyzed strategy provides a consistent and reproducible means to conjugate fluorescent dyes, cytotoxins, or "click" chemistry handles to meditope-enabled mAbs (memAbs) and memFabs. More importantly, we demonstrate this covalent functionalization is achievable using natural amino acids only, opening up the opportunity to genetically encode cysteine meditope "tags" to biologics. As proof of principle, genetically encoded, cysteine meditope tags were added to the N- and/or C-termini of fluorescent proteins, nanobodies, and affibodies, each expressed in bacteria, purified to homogeneity, and efficiently conjugated to different memAbs and meFabs. We further show that multiple T-cell and Her2-targeting bispecific molecules using this strategy potently activate T-cell signaling pathways in vitro. Finally, the resulting products are highly stable as evidenced by serum stability assays (>14 d at 37 °C) and in vivo imaging of tumor xenographs. Collectively, the platform offers the opportunity to build and exchange an array of functional moieties, including protein biologics, among any cysteine memAb or Fab to rapidly create, test, and optimize stable, multifunctional biologics.