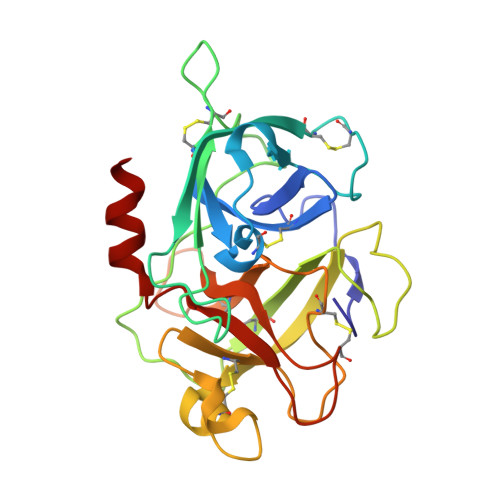
Structures of human plasma beta-factor XIIa cocrystallized with potent inhibitors.
Dementiev, A., Silva, A., Yee, C., Li, Z., Flavin, M.T., Sham, H., Partridge, J.R.(2018) Blood Adv 2: 549-558
- PubMed: 29519898
- DOI: https://doi.org/10.1182/bloodadvances.2018016337
- Primary Citation of Related Structures:
6B74, 6B77 - PubMed Abstract:
Activated factor XIIa (FXIIa) is a serine protease that has received a great deal of interest in recent years as a potential target for the development of new antithrombotics. Despite the strong interest in obtaining structural information, only the structure of the FXIIa catalytic domain in its zymogen conformation is available. In this work, reproducible experimental conditions found for the crystallization of human plasma β-FXIIa and crystal growth optimization have led to determination of the first structure of the active form of the enzyme. Two crystal structures of human plasma β-FXIIa complexed with small molecule inhibitors are presented herein. The first is the noncovalent inhibitor benzamidine. The second is an aminoisoquinoline containing a boronic acid-reactive group that targets the catalytic serine. Both benzamidine and the aminoisoquinoline bind in a canonical fashion typical of synthetic serine protease inhibitors, and the protease domain adopts a typical chymotrypsin-like serine protease active conformation. This novel structural data explains the basis of the FXII activation, provides insights into the enzymatic properties of β-FXIIa, and is a great aid toward the further design of protease inhibitors for human FXIIa.
- Shamrock Structures, LLC, Woodridge, IL; and.
Organizational Affiliation: