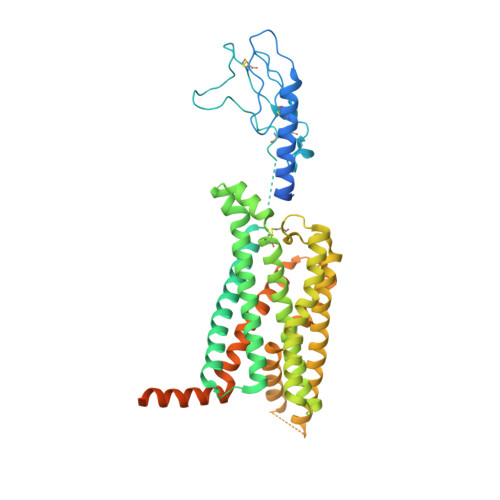
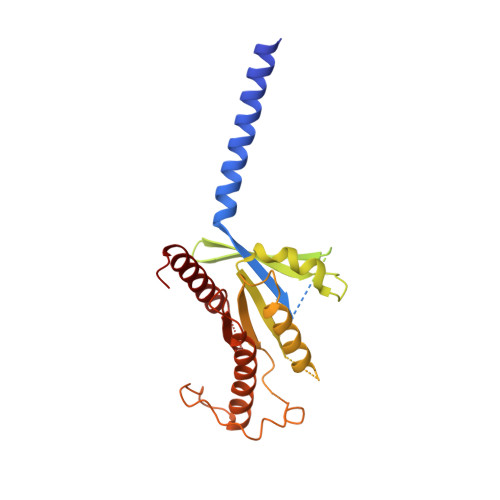
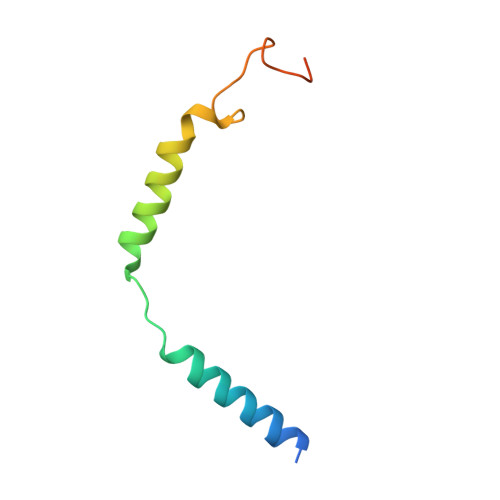
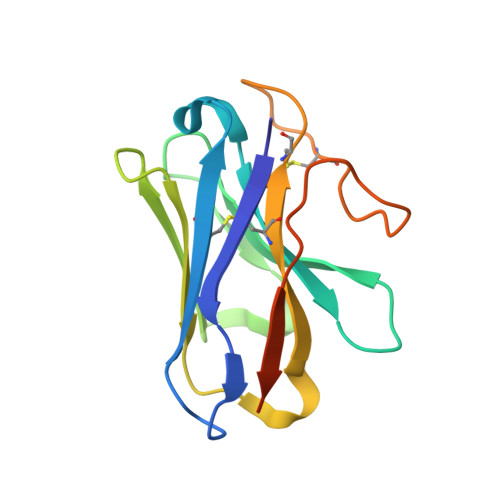
Phase-plate cryo-EM structure of a biased agonist-bound human GLP-1 receptor-Gs complex.
Liang, Y.L., Khoshouei, M., Glukhova, A., Furness, S.G.B., Zhao, P., Clydesdale, L., Koole, C., Truong, T.T., Thal, D.M., Lei, S., Radjainia, M., Danev, R., Baumeister, W., Wang, M.W., Miller, L.J., Christopoulos, A., Sexton, P.M., Wootten, D.(2018) Nature 555: 121-125
- PubMed: 29466332
- DOI: https://doi.org/10.1038/nature25773
- Primary Citation of Related Structures:
6B3J - PubMed Abstract:
The class B glucagon-like peptide-1 (GLP-1) G protein-coupled receptor is a major target for the treatment of type 2 diabetes and obesity. Endogenous and mimetic GLP-1 peptides exhibit biased agonism-a difference in functional selectivity-that may provide improved therapeutic outcomes. Here we describe the structure of the human GLP-1 receptor in complex with the G protein-biased peptide exendin-P5 and a Gα s heterotrimer, determined at a global resolution of 3.3 Å. At the extracellular surface, the organization of extracellular loop 3 and proximal transmembrane segments differs between our exendin-P5-bound structure and previous GLP-1-bound GLP-1 receptor structure. At the intracellular face, there was a six-degree difference in the angle of the Gαs-α5 helix engagement between structures, which was propagated across the G protein heterotrimer. In addition, the structures differed in the rate and extent of conformational reorganization of the Gα s protein. Our structure provides insights into the molecular basis of biased agonism.
- Drug Discovery Biology and Department of Pharmacology, Monash Institute of Pharmaceutical Sciences, Monash University, Parkville 3052, Victoria, Australia.
Organizational Affiliation: