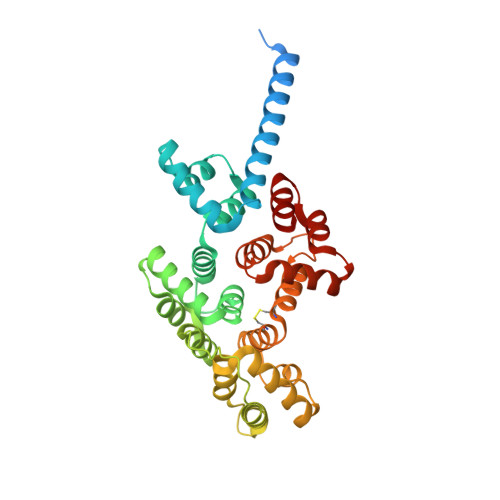
An alternative N-terminal fold of the intestine-specific annexin A13a induces dimerization and regulates membrane-binding.
McCulloch, K.M., Yamakawa, I., Shifrin Jr., D.A., McConnell, R.E., Foegeding, N.J., Singh, P.K., Mao, S., Tyska, M.J., Iverson, T.M.(2019) J Biological Chem 294: 3454-3463
- PubMed: 30610115
- DOI: https://doi.org/10.1074/jbc.RA118.004571
- Primary Citation of Related Structures:
6B3I - PubMed Abstract:
Annexin proteins function as Ca 2+ -dependent regulators of membrane trafficking and repair that may also modulate membrane curvature. Here, using high-resolution confocal imaging, we report that the intestine-specific annexin A13 (ANX A13) localizes to the tips of intestinal microvilli and determined the crystal structure of the ANX A13a isoform to 2.6 Å resolution. The structure revealed that the N terminus exhibits an alternative fold that converts the first two helices and the associated helix-loop-helix motif into a continuous α-helix, as stabilized by a domain-swapped dimer. We also found that the dimer is present in solution and partially occludes the membrane-binding surfaces of annexin, suggesting that dimerization may function as a means for regulating membrane binding. Accordingly, as revealed by in vitro binding and cellular localization assays, ANX A13a variants that favor a monomeric state exhibited increased membrane association relative to variants that favor the dimeric form. Together, our findings support a mechanism for how the association of the ANX A13a isoform with the membrane is regulated.
- From the Departments of Pharmacology.
Organizational Affiliation: