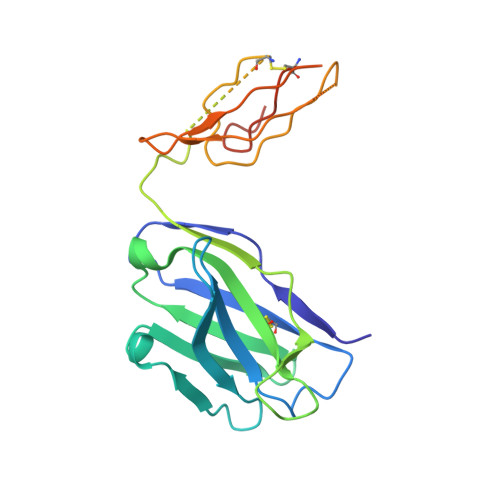
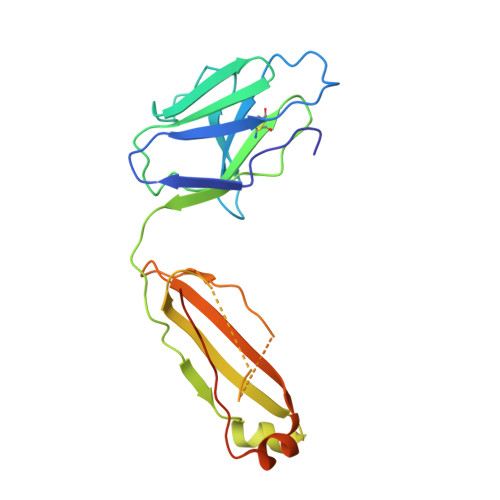
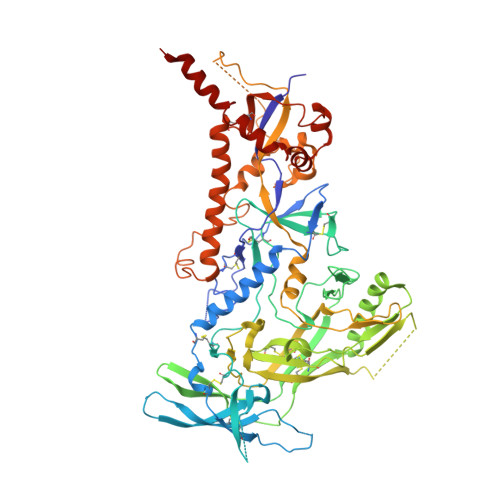
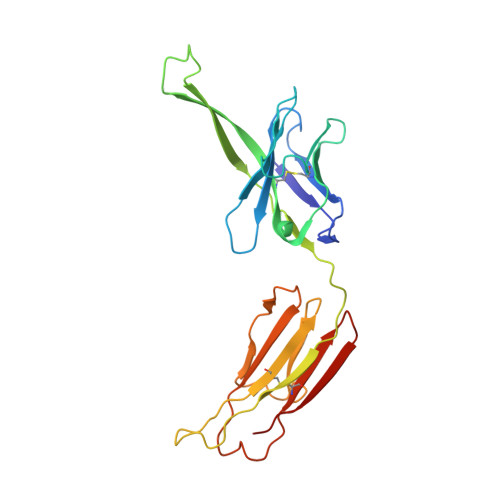
Structure of a cleavage-independent HIV Env recapitulates the glycoprotein architecture of the native cleaved trimer.
Sarkar, A., Bale, S., Behrens, A.J., Kumar, S., Sharma, S.K., de Val, N., Pallesen, J., Irimia, A., Diwanji, D.C., Stanfield, R.L., Ward, A.B., Crispin, M., Wyatt, R.T., Wilson, I.A.(2018) Nat Commun 9: 1956-1956
- PubMed: 29769533
- DOI: https://doi.org/10.1038/s41467-018-04272-y
- Primary Citation of Related Structures:
6AVN, 6B0N - PubMed Abstract:
Furin cleavage of the HIV envelope glycoprotein is an essential step for cell entry that enables formation of well-folded, native-like glycosylated trimers, releases constraints on the fusion peptide, and limits enzymatic processing of the N-glycan shield. Here, we show that a cleavage-independent, stabilized, soluble Env trimer mimic (BG505 NFL.664) exhibits a "closed-form", native-like, prefusion conformation akin to furin-cleaved Env trimers. The crystal structure of BG505 NFL.664 at 3.39 Å resolution with two potent bNAbs also identifies the full epitopes of PGV19 and PGT122 that target the receptor binding site and N332 supersite, respectively. Quantitative site-specific analysis of the glycan shield reveals that native-like glycan processing is maintained despite furin-independent maturation in the secretory pathway. Thus, cleavage-independent NFL Env trimers exhibit quaternary protein and carbohydrate structures similar to the native viral spike that further validate their potential as vaccine immunogen candidates.
- IAVI Neutralizing Antibody Center, The Scripps Research Institute, La Jolla, CA, 92037, USA.
Organizational Affiliation: