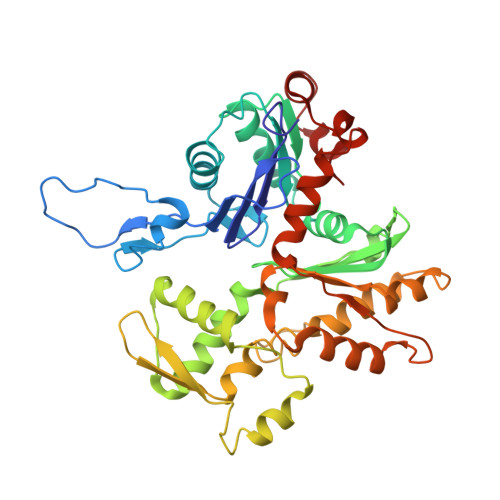
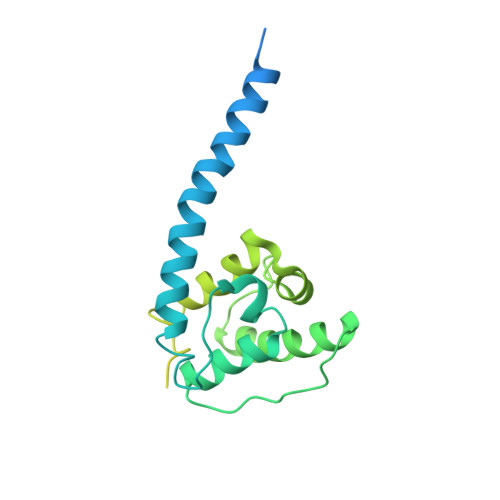
Structural basis for high-affinity actin binding revealed by a beta-III-spectrin SCA5 missense mutation.
Avery, A.W., Fealey, M.E., Wang, F., Orlova, A., Thompson, A.R., Thomas, D.D., Hays, T.S., Egelman, E.H.(2017) Nat Commun 8: 1350-1350
- PubMed: 29116080
- DOI: https://doi.org/10.1038/s41467-017-01367-w
- Primary Citation of Related Structures:
6ANU - PubMed Abstract:
Spinocerebellar ataxia type 5 (SCA5) is a neurodegenerative disease caused by mutations in the cytoskeletal protein β-III-spectrin. Previously, a SCA5 mutation resulting in a leucine-to-proline substitution (L253P) in the actin-binding domain (ABD) was shown to cause a 1000-fold increase in actin-binding affinity. However, the structural basis for this increase is unknown. Here, we report a 6.9 Å cryo-EM structure of F-actin complexed with the L253P ABD. This structure, along with co-sedimentation and pulsed-EPR measurements, demonstrates that high-affinity binding caused by the CH2-localized mutation is due to opening of the two CH domains. This enables CH1 to bind actin aided by an unstructured N-terminal region that becomes α-helical upon binding. This helix is required for association with actin as truncation eliminates binding. Collectively, these results shed light on the mechanism by which β-III-spectrin, and likely similar actin-binding proteins, interact with actin, and how this mechanism can be perturbed to cause disease.
- Department of Genetics, Cell Biology and Development, University of Minnesota, Minneapolis, MN, 55455, USA.
Organizational Affiliation: