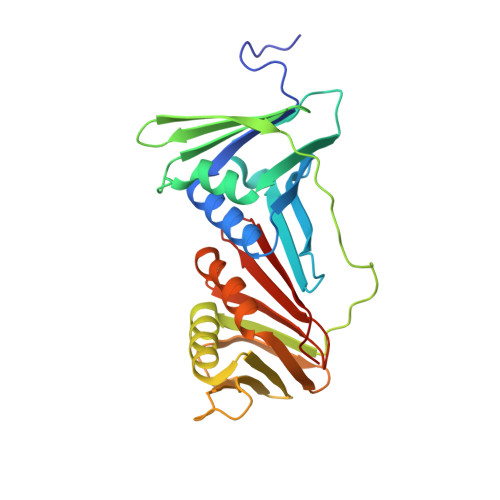
X-ray crystal structure of proliferating cell nuclear antigen 1 from Aeropyrum pernix.
Yamauchi, T., Kikuchi, M., Iizuka, Y., Tsunoda, M.(2024) Acta Crystallogr F Struct Biol Commun 80: 294-301
- PubMed: 39382846
- DOI: https://doi.org/10.1107/S2053230X24009518
- Primary Citation of Related Structures:
6AIG - PubMed Abstract:
Proliferating cell nuclear antigen (PCNA) plays a critical role in DNA replication by enhancing the activity of various proteins involved in replication. In this study, the crystal structure of ApePCNA1, one of three PCNAs from the thermophilic archaeon Aeropyrum pernix, was elucidated. ApePCNA1 was cloned and expressed in Escherichia coli and the protein was purified and crystallized. The resulting crystal structure determined at 2.00 Å resolution revealed that ApePCNA1 does not form a trimeric ring, unlike PCNAs from other domains of life. It has unique structural features, including a long interdomain-connecting loop and a PIP-box-like sequence at the N-terminus, indicating potential interactions with other proteins. These findings provide insights into the functional mechanisms of PCNAs in archaea and their evolutionary conservation across different domains of life. A modified medium and protocol were used to express recombinant protein containing the lac operon. The expression of the target protein increased and the total incubation time decreased when using this system compared with those of previous expression protocols.
- Graduate School of Life Science and Technology, Iryo Sosei University, Iwaki, Fukushima, Japan.
Organizational Affiliation: