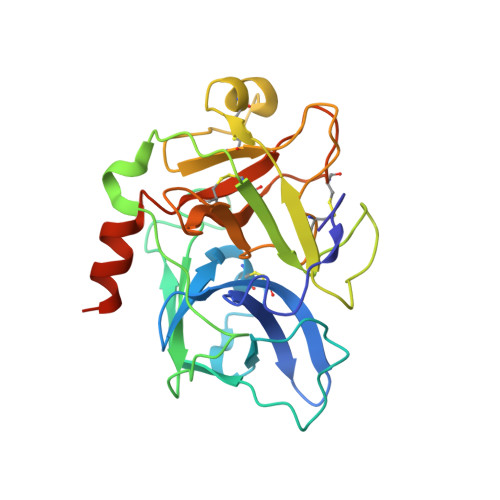
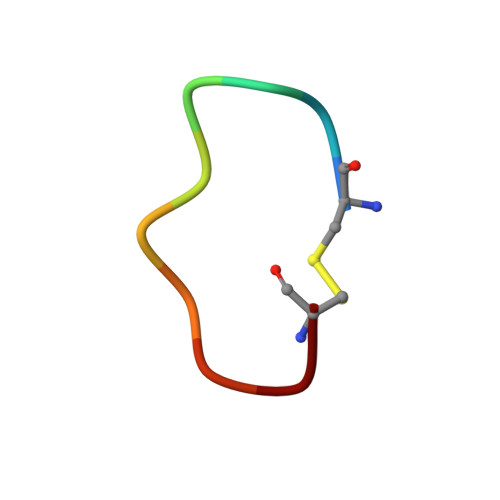
Crystal structure of plasma kallikrein reveals the unusual flexibility of the S1 pocket triggered by Glu217.
Xu, M., Chen, Y., Xu, P., Andreasen, P.A., Jiang, L., Li, J., Huang, M.(2018) FEBS Lett 592: 2658-2667
- PubMed: 30019481
- DOI: https://doi.org/10.1002/1873-3468.13191
- Primary Citation of Related Structures:
6A8O - PubMed Abstract:
Serine proteases play important roles in numerous physiological and pathophysiological processes. Moreover, serine proteases are classical subjects for studies of catalytic and inhibitory mechanisms of enzymes. Here, we determined the crystal structures of a serine protease, murine plasma kallikrein (mPK), and its complex with a peptidic inhibitor. Although mPK in the complex adopts a canonical protease structure, the apo-mPK exhibits a previously unobserved structural feature: the entrance of the intact S1 pocket is blocked by Glu217. In addition, molecular dynamics simulations and functional assays support the flexibility of Glu217 and suggest that this flexibility plays a role in regulating the activity of serine proteases. EC: 3.4.21.34.
- College of Chemistry, Fuzhou University, China.
Organizational Affiliation: