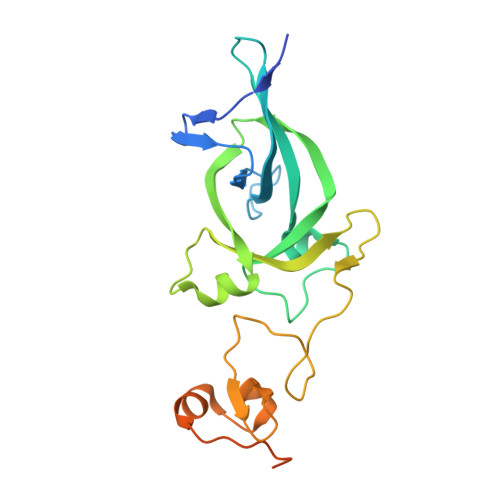
Dual recognition of H3K4me3 and H3K27me3 by a plant histone reader SHL.
Qian, S., Lv, X., Scheid, R.N., Lu, L., Yang, Z., Chen, W., Liu, R., Boersma, M.D., Denu, J.M., Zhong, X., Du, J.(2018) Nat Commun 9: 2425-2425
- PubMed: 29930355
- DOI: https://doi.org/10.1038/s41467-018-04836-y
- Primary Citation of Related Structures:
5ZNP, 5ZNR - PubMed Abstract:
The ability of a cell to dynamically switch its chromatin between different functional states constitutes a key mechanism regulating gene expression. Histone mark "readers" display distinct binding specificity to different histone modifications and play critical roles in regulating chromatin states. Here, we show a plant-specific histone reader SHORT LIFE (SHL) capable of recognizing both H3K27me3 and H3K4me3 via its bromo-adjacent homology (BAH) and plant homeodomain (PHD) domains, respectively. Detailed biochemical and structural studies suggest a binding mechanism that is mutually exclusive for either H3K4me3 or H3K27me3. Furthermore, we show a genome-wide co-localization of SHL with H3K27me3 and H3K4me3, and that BAH-H3K27me3 and PHD-H3K4me3 interactions are important for SHL-mediated floral repression. Together, our study establishes BAH-PHD cassette as a dual histone methyl-lysine binding module that is distinct from others in recognizing both active and repressive histone marks.
- Laboratory of Genetics, University of Wisconsin-Madison, Madison, WI, 53706, USA.
Organizational Affiliation: