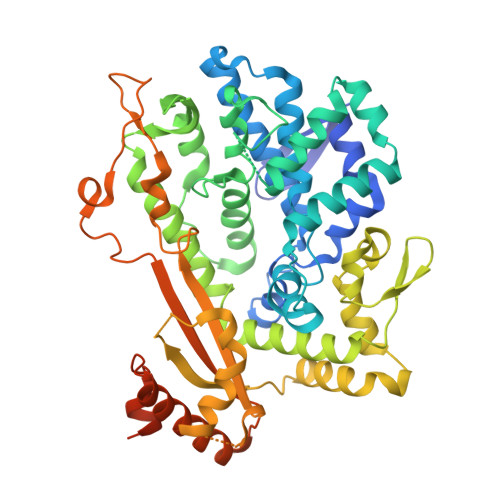
Structural characterization and directed modification of Sus scrofa SAMHD1 reveal the mechanism underlying deoxynucleotide regulation.
Kong, J., Wang, M.M., He, S.Y., Peng, X., Qin, X.H.(2019) FEBS J 286: 3844-3857
- PubMed: 31152619
- DOI: https://doi.org/10.1111/febs.14943
- Primary Citation of Related Structures:
5YHW - PubMed Abstract:
Sterile α-motif/histidine-aspartate domain-containing protein 1 (SAMHD1) is an intrinsic antiviral restriction factor known to play a vital role in preventing multiple viral infections and in the control of the cellular deoxynucleoside triphosphate (dNTP) pool. Human and mouse SAMHD1 have both been extensively studied; however, our knowledge on porcine SAMHD1 is limited. Here, we report our findings from comprehensive structural and functional studies on porcine SAMHD1. We determined the crystal structure of porcine SAMHD1 and showed that it forms a symmetric tetramer. Moreover, we modified the deoxynucleotide triphosphohydrolase (dNTPase) activity of SAMHD1 by site-directed mutagenesis based on the crystal structure, and obtained an artificial dimeric enzyme possessing high dNTPase activity. Taken together, our results define the mechanism underlying dNTP regulation and provide a deeper understanding of the regulation of porcine SAMHD1 functions. Directed modification of key residues based on the protein structure enhances the activity of the enzyme, which will be beneficial in the search for new antiviral strategies and for future translational applications.
- School of Chemical Engineering and Technology, Tianjin University, China.
Organizational Affiliation: