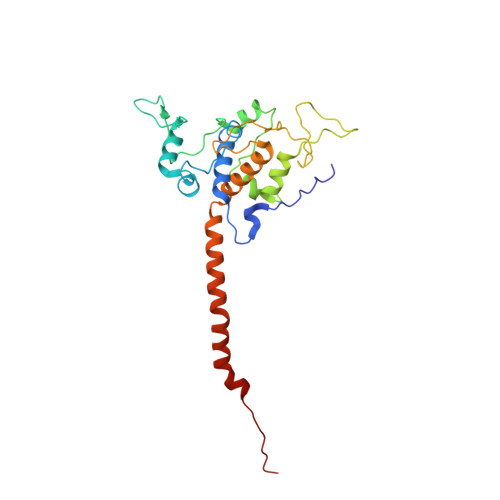
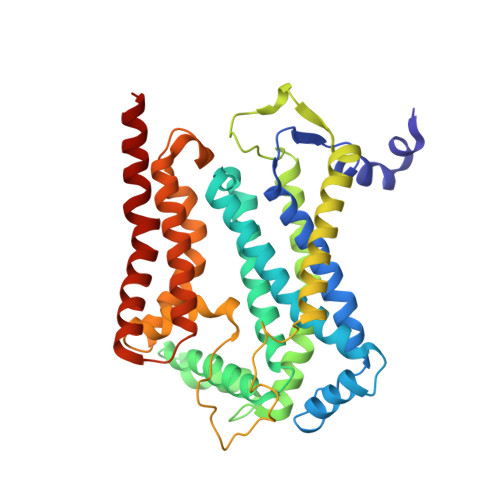
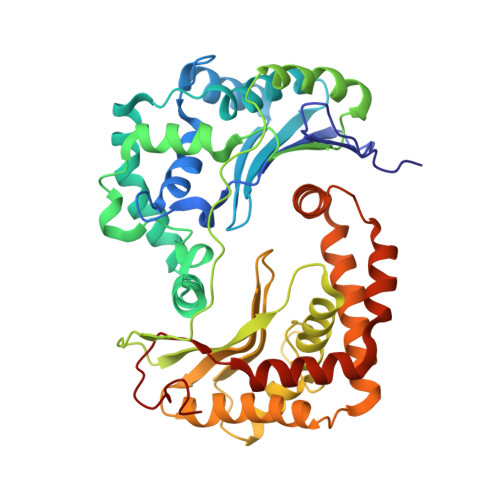
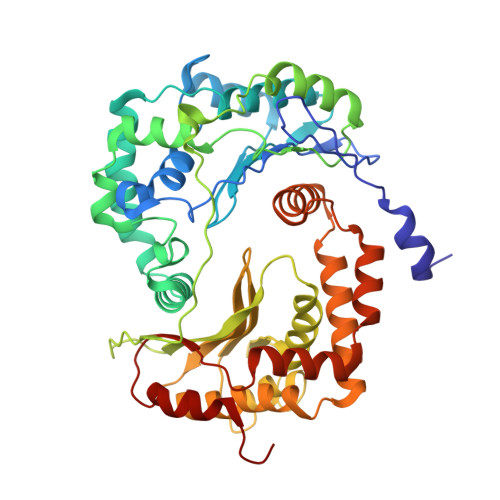
Architecture of Human Mitochondrial Respiratory Megacomplex I2III2IV2.
Guo, R., Zong, S., Wu, M., Gu, J., Yang, M.(2017) Cell 170: 1247-1257.e12
- PubMed: 28844695
- DOI: https://doi.org/10.1016/j.cell.2017.07.050
- Primary Citation of Related Structures:
5XTB, 5XTC, 5XTD, 5XTE, 5XTH, 5XTI - PubMed Abstract:
The respiratory megacomplex represents the highest-order assembly of respiratory chain complexes, and it allows mitochondria to respond to energy-requiring conditions. To understand its architecture, we examined the human respiratory chain megacomplex-I 2 III 2 IV 2 (MCI 2 III 2 IV 2 ) with 140 subunits and a subset of associated cofactors using cryo-electron microscopy. The MCI 2 III 2 IV 2 forms a circular structure with the dimeric CIII located in the center, where it is surrounded by two copies each of CI and CIV. Two cytochrome c (Cyt.c) molecules are positioned to accept electrons on the surface of the c 1 state CIII dimer. Analyses indicate that CII could insert into the gaps between CI and CIV to form a closed ring, which we termed the electron transport chain supercomplex. The structure not only reveals the precise assignment of individual subunits of human CI and CIII, but also enables future in-depth analysis of the electron transport chain as a whole.
- Ministry of Education Key Laboratory of Protein Science, Tsinghua-Peking Joint Center for Life Sciences, Beijing Advanced Innovation Center for Structural Biology, School of Life Sciences, Tsinghua University, 100084 Beijing, China.
Organizational Affiliation: