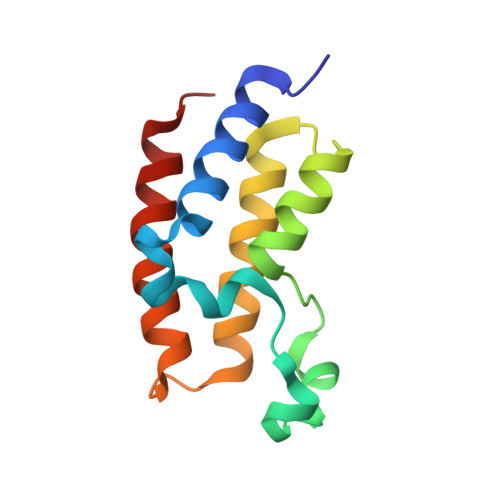
Distinct Roles of Brd2 and Brd4 in Potentiating the Transcriptional Program for Th17 Cell Differentiation.
Cheung, K.L., Zhang, F., Jaganathan, A., Sharma, R., Zhang, Q., Konuma, T., Shen, T., Lee, J.Y., Ren, C., Chen, C.H., Lu, G., Olson, M.R., Zhang, W., Kaplan, M.H., Littman, D.R., Walsh, M.J., Xiong, H., Zeng, L., Zhou, M.M.(2017) Mol Cell 65: 1068-1080.e5
- PubMed: 28262505
- DOI: https://doi.org/10.1016/j.molcel.2016.12.022
- Primary Citation of Related Structures:
5U5S - PubMed Abstract:
The BET proteins are major transcriptional regulators and have emerged as new drug targets, but their functional distinction has remained elusive. In this study, we report that the BET family members Brd2 and Brd4 exert distinct genomic functions at genes whose transcription they co-regulate during mouse T helper 17 (Th17) cell differentiation. Brd2 is associated with the chromatin insulator CTCF and the cohesin complex to support cis-regulatory enhancer assembly for gene transcriptional activation. In this context, Brd2 binds the transcription factor Stat3 in an acetylation-sensitive manner and facilitates Stat3 recruitment to active enhancers occupied with transcription factors Irf4 and Batf. In parallel, Brd4 temporally controls RNA polymerase II (Pol II) processivity during transcription elongation through cyclin T1 and Cdk9 recruitment and Pol II Ser2 phosphorylation. Collectively, our study uncovers both separate and interdependent Brd2 and Brd4 functions in potentiating the genetic program required for Th17 cell development and adaptive immunity.
- Department of Pharmacological Sciences, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA.
Organizational Affiliation: