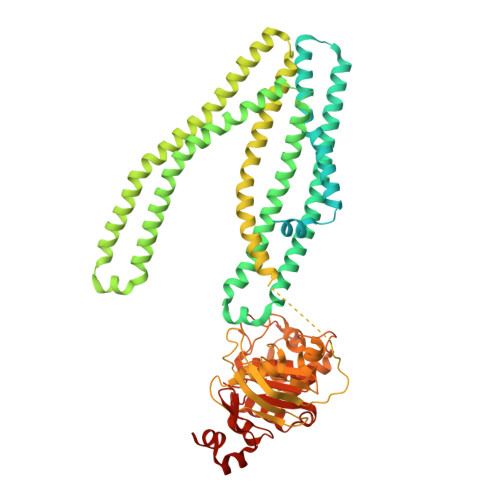
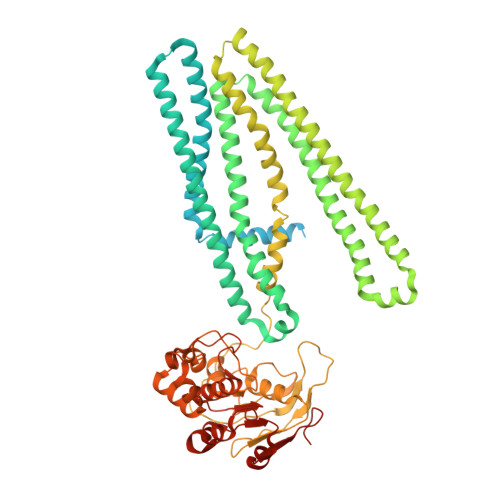
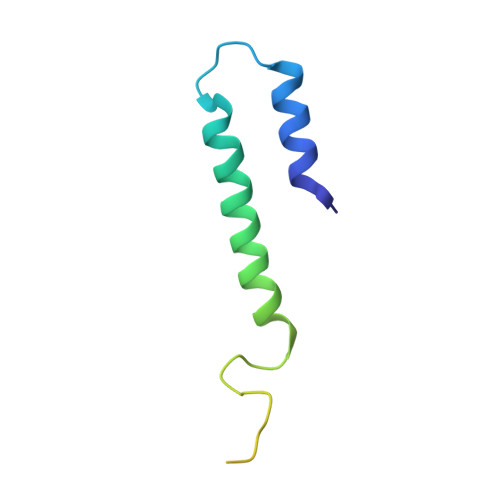
Structure of the transporter associated with antigen processing trapped by herpes simplex virus.
Oldham, M.L., Grigorieff, N., Chen, J.(2016) Elife 5
- PubMed: 27935481
- DOI: https://doi.org/10.7554/eLife.21829
- Primary Citation of Related Structures:
5U1D - PubMed Abstract:
The transporter associated with antigen processing (TAP) is an ATP-binding cassette (ABC) transporter essential to cellular immunity against viral infection. Some persistent viruses have evolved strategies to inhibit TAP so that they may go undetected by the immune system. The herpes simplex virus for example evades immune surveillance by blocking peptide transport with a small viral protein ICP47. In this study, we determined the structure of human TAP bound to ICP47 by electron cryo-microscopy (cryo-EM) to 4.0 Å. The structure shows that ICP47 traps TAP in an inactive conformation distinct from the normal transport cycle. The specificity and potency of ICP47 inhibition result from contacts between the tip of the helical hairpin and the apex of the transmembrane cavity. This work provides a clear molecular description of immune evasion by a persistent virus. It also establishes the molecular structure of TAP to facilitate mechanistic studies of the antigen presentation process.
- Howard Hughes Medical Institute, The Rockefeller University, New York, United States.
Organizational Affiliation: